Early adversity changes the economic conditions of mouse structural brain network organization
- PMID: 37607894
- PMCID: PMC10505050
- DOI: 10.1002/dev.22405
Early adversity changes the economic conditions of mouse structural brain network organization
Abstract
Early adversity can change educational, cognitive, and mental health outcomes. However, the neural processes through which early adversity exerts these effects remain largely unknown. We used generative network modeling of the mouse connectome to test whether unpredictable postnatal stress shifts the constraints that govern the organization of the structural connectome. A model that trades off the wiring cost of long-distance connections with topological homophily (i.e., links between regions with shared neighbors) generated simulations that successfully replicate the rodent connectome. The imposition of early life adversity shifted the best-performing parameter combinations toward zero, heightening the stochastic nature of the generative process. Put simply, unpredictable postnatal stress changes the economic constraints that reproduce rodent connectome organization, introducing greater randomness into the development of the simulations. While this change may constrain the development of cognitive abilities, it could also reflect an adaptive mechanism that facilitates effective responses to future challenges.
Keywords: brain organization; early adversity; generative models; graph theory; structural connectome; unpredictable stress.
© 2023 Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
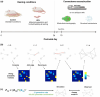

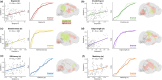


References
-
- Akarca, D. , Dunn, A. W. E. , Hornauer, P. J. , Ronchi, S. , Fiscella, M. , Wang, C. , Terrigno, M. , Jagasia, R. , Vértes, P. E. , Mierau, S. B. , Paulsen, O. , Eglen, S. J. , Hierlemann, A. , Astle, D. E. , & Schröter, M. (2022). Homophilic wiring principles underpin neuronal network topology in vitro. bioRxiv, 10.1101/2022.03.09.483605 - DOI
-
- Akarca, D. , Vértes, P. E. , Bullmore, E. T. , Baker, K. , Gathercole, S. E. , Holmes, J. , Kievit, R. A. , Manly, T. , Bathelt, J. , Bennett, M. , Bignardi, G. , Bishop, S. , Bottacin, E. , Bridge, L. , Brkic, D. , Bryant, A. , Butterfield, S. , Byrne, E. M. , Crickmore, G. , … Astle, D. E. (2021). A generative network model of neurodevelopmental diversity in structural brain organization. Nature Communications, 12(1), 1–18. 10.1038/s41467-021-24430-z - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

