TUG1-mediated R-loop resolution at microsatellite loci as a prerequisite for cancer cell proliferation
- PMID: 37607907
- PMCID: PMC10444773
- DOI: 10.1038/s41467-023-40243-8
TUG1-mediated R-loop resolution at microsatellite loci as a prerequisite for cancer cell proliferation
Abstract
Oncogene-induced DNA replication stress (RS) and consequent pathogenic R-loop formation are known to impede S phase progression. Nonetheless, cancer cells continuously proliferate under such high-stressed conditions through incompletely understood mechanisms. Here, we report taurine upregulated gene 1 (TUG1) long noncoding RNA (lncRNA), which is highly expressed in many types of cancers, as an important regulator of intrinsic R-loop in cancer cells. Under RS conditions, TUG1 is rapidly upregulated via activation of the ATR-CHK1 signaling pathway, interacts with RPA and DHX9, and engages in resolving R-loops at certain loci, particularly at the CA repeat microsatellite loci. Depletion of TUG1 leads to overabundant R-loops and enhanced RS, leading to substantial inhibition of tumor growth. Our data reveal a role of TUG1 as molecule important for resolving R-loop accumulation in cancer cells and suggest targeting TUG1 as a potent therapeutic approach for cancer treatment.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

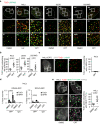
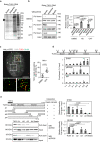
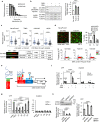
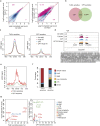

References
-
- Kotsantis P, Petermann E, Boulton SJ. Mechanisms of oncogene-induced replication stress: jigsaw falling into place. Cancer Discov. 2018;8:537–555. doi: 10.1158/2159-8290.CD-17-1461. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

