A20 promotes colorectal cancer immune evasion by upregulating STC1 expression to block "eat-me" signal
- PMID: 37607946
- PMCID: PMC10444827
- DOI: 10.1038/s41392-023-01545-x
A20 promotes colorectal cancer immune evasion by upregulating STC1 expression to block "eat-me" signal
Abstract
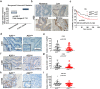
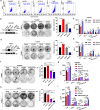
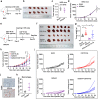
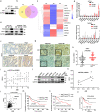
Immune checkpoint inhibitors (ICIs) have induced durable clinical responses in a subset of patients with colorectal cancer (CRC). However, the dis-satisfactory response rate and the lack of appropriate biomarkers for selecting suitable patients to be treated with ICIs pose a major challenge to current immunotherapies. Inflammation-related molecule A20 is closely related to cancer immune response, but the effect of A20 on "eat-me" signal and immunotherapy efficacy remains elusive. We found that A20 downregulation prominently improved the antitumor immune response and the efficacy of PD-1 inhibitor in CRC in vitro and in vivo. Higher A20 expression was associated with less infiltration of immune cells including CD3 (+), CD8 (+) T cells and macrophages in CRC tissues and also poorer prognosis. Gain- and loss-A20 functional studies proved that A20 could decrease the "eat-me" signal calreticulin (CRT) protein on cell membrane translocation via upregulating stanniocalcin 1 (STC1), binding to CRT and detaining in mitochondria. Mechanistically, A20 inhibited GSK3β phosphorylating STC1 at Thr86 to slow down the degradation of STC1 protein. Our findings reveal a new crosstalk between inflammatory molecule A20 and "eat-me" signal in CRC, which may represent a novel predictive biomarker for selecting CRC patients most likely to benefit from ICI therapy.
© 2023. West China Hospital, Sichuan University.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Stanniocalcin 1 is a phagocytosis checkpoint driving tumor immune resistance.Cancer Cell. 2021 Apr 12;39(4):480-493.e6. doi: 10.1016/j.ccell.2020.12.023. Epub 2021 Jan 28. Cancer Cell. 2021. PMID: 33513345 Free PMC article.
-
A20 regulates the therapeutic effect of anti-PD-1 immunotherapy in melanoma.J Immunother Cancer. 2020 Dec;8(2):e001866. doi: 10.1136/jitc-2020-001866. J Immunother Cancer. 2020. PMID: 33298620 Free PMC article.
-
Blocking IL-17A enhances tumor response to anti-PD-1 immunotherapy in microsatellite stable colorectal cancer.J Immunother Cancer. 2021 Jan;9(1):e001895. doi: 10.1136/jitc-2020-001895. J Immunother Cancer. 2021. PMID: 33462141 Free PMC article.
-
Relationships Between Immune Landscapes, Genetic Subtypes and Responses to Immunotherapy in Colorectal Cancer.Front Immunol. 2020 Mar 6;11:369. doi: 10.3389/fimmu.2020.00369. eCollection 2020. Front Immunol. 2020. PMID: 32210966 Free PMC article. Review.
-
T-cell immunoglobulin and ITIM domain, as a potential immune checkpoint target for immunotherapy of colorectal cancer.IUBMB Life. 2021 May;73(5):726-738. doi: 10.1002/iub.2461. Epub 2021 Mar 30. IUBMB Life. 2021. PMID: 33686787 Review.
Cited by
-
Deubiquitinases as novel therapeutic targets for diseases.MedComm (2020). 2024 Dec 13;5(12):e70036. doi: 10.1002/mco2.70036. eCollection 2024 Dec. MedComm (2020). 2024. PMID: 39678489 Free PMC article. Review.
-
Integrative analysis of recurrence related gene signature and STC1 in colorectal cancer proliferation and metastasis.J Cancer. 2024 Oct 28;15(20):6724-6739. doi: 10.7150/jca.102605. eCollection 2024. J Cancer. 2024. PMID: 39668832 Free PMC article.
-
Dissecting the dual role of OTU family proteins in tumor progression and immune escape.Front Immunol. 2025 May 21;16:1544341. doi: 10.3389/fimmu.2025.1544341. eCollection 2025. Front Immunol. 2025. PMID: 40469292 Free PMC article. Review.
-
CVD Atlas: a multi-omics database of cardiovascular disease.Nucleic Acids Res. 2025 Jan 6;53(D1):D1348-D1355. doi: 10.1093/nar/gkae848. Nucleic Acids Res. 2025. PMID: 39351894 Free PMC article.
-
Advances in cancer immunotherapy: historical perspectives, current developments, and future directions.Mol Cancer. 2025 May 7;24(1):136. doi: 10.1186/s12943-025-02305-x. Mol Cancer. 2025. PMID: 40336045 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

