Identifying COVID-19 cases and extracting patient reported symptoms from Reddit using natural language processing
- PMID: 37607963
- PMCID: PMC10444846
- DOI: 10.1038/s41598-023-39986-7
Identifying COVID-19 cases and extracting patient reported symptoms from Reddit using natural language processing
Abstract
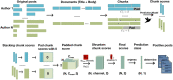
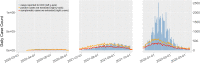
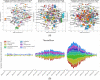
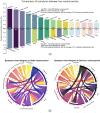
We used social media data from "covid19positive" subreddit, from 03/2020 to 03/2022 to identify COVID-19 cases and extract their reported symptoms automatically using natural language processing (NLP). We trained a Bidirectional Encoder Representations from Transformers classification model with chunking to identify COVID-19 cases; also, we developed a novel QuadArm model, which incorporates Question-answering, dual-corpus expansion, Adaptive rotation clustering, and mapping, to extract symptoms. Our classification model achieved a 91.2% accuracy for the early period (03/2020-05/2020) and was applied to the Delta (07/2021-09/2021) and Omicron (12/2021-03/2022) periods for case identification. We identified 310, 8794, and 12,094 COVID-positive authors in the three periods, respectively. The top five common symptoms extracted in the early period were coughing (57%), fever (55%), loss of sense of smell (41%), headache (40%), and sore throat (40%). During the Delta period, these symptoms remained as the top five symptoms with percent authors reporting symptoms reduced to half or fewer than the early period. During the Omicron period, loss of sense of smell was reported less while sore throat was reported more. Our study demonstrated that NLP can be used to identify COVID-19 cases accurately and extracted symptoms efficiently.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Using Social Media to Help Understand Patient-Reported Health Outcomes of Post-COVID-19 Condition: Natural Language Processing Approach.J Med Internet Res. 2023 Sep 19;25:e45767. doi: 10.2196/45767. J Med Internet Res. 2023. PMID: 37725432 Free PMC article.
-
COVID-19 Symptoms and Duration of Rapid Antigen Test Positivity at a Community Testing and Surveillance Site During Pre-Delta, Delta, and Omicron BA.1 Periods.JAMA Netw Open. 2022 Oct 3;5(10):e2235844. doi: 10.1001/jamanetworkopen.2022.35844. JAMA Netw Open. 2022. PMID: 36215069 Free PMC article.
-
Natural Language Processing for Improved Characterization of COVID-19 Symptoms: Observational Study of 350,000 Patients in a Large Integrated Health Care System.JMIR Public Health Surveill. 2022 Dec 30;8(12):e41529. doi: 10.2196/41529. JMIR Public Health Surveill. 2022. PMID: 36446133 Free PMC article.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2021 Feb 23;2(2):CD013665. doi: 10.1002/14651858.CD013665.pub2. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 20;5:CD013665. doi: 10.1002/14651858.CD013665.pub3. PMID: 33620086 Free PMC article. Updated.
-
Natural language processing of symptoms documented in free-text narratives of electronic health records: a systematic review.J Am Med Inform Assoc. 2019 Apr 1;26(4):364-379. doi: 10.1093/jamia/ocy173. J Am Med Inform Assoc. 2019. PMID: 30726935 Free PMC article.
Cited by
-
BERT-based language model for accurate drug adverse event extraction from social media: implementation, evaluation, and contributions to pharmacovigilance practices.Front Public Health. 2024 Apr 23;12:1392180. doi: 10.3389/fpubh.2024.1392180. eCollection 2024. Front Public Health. 2024. PMID: 38716250 Free PMC article.
-
Pharmacovigilance in the digital age: gaining insight from social media data.Exp Biol Med (Maywood). 2025 May 27;250:10555. doi: 10.3389/ebm.2025.10555. eCollection 2025. Exp Biol Med (Maywood). 2025. PMID: 40495881 Free PMC article. Review.
-
LLM enabled classification of patient self-reported symptoms and needs in health systems across the USA.NPJ Digit Med. 2025 Jul 1;8(1):390. doi: 10.1038/s41746-025-01779-9. NPJ Digit Med. 2025. PMID: 40595018 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

