Induction of open-form bile canaliculus formation by hepatocytes for evaluation of biliary drug excretion
- PMID: 37608051
- PMCID: PMC10444810
- DOI: 10.1038/s42003-023-05216-z
Induction of open-form bile canaliculus formation by hepatocytes for evaluation of biliary drug excretion
Abstract
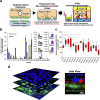
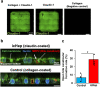
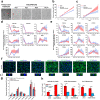
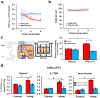
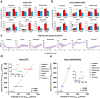
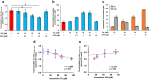
Biliary excretion is a major drug elimination pathway that affects their efficacy and safety. The currently available in vitro sandwich-cultured hepatocyte method is cumbersome because drugs accumulate in the closed bile canalicular lumen formed between hepatocytes and their amounts cannot be mealsured directly. This study proposes a hepatocyte culture model for the rapid evaluation of drug biliary excretion using permeation assays. When hepatocytes are cultured on a permeable support coated with the cell adhesion protein claudins, an open-form bile canalicular lumen is formed at the surface of the permeable support. Upon application to the basolateral (blood) side, drugs appear on the bile canalicular side. The biliary excretion clearance of several drugs, as estimated from the obtained permeabilities, correlates well with the reported in vivo biliary excretion clearance in humans. Thus, the established model is useful for applications in the efficient evaluation of biliary excretion during drug discovery and development.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Construction of extended and functional bile canaliculi using long-term sandwich-cultured cryopreserved human hepatocytes and the application of hepatocytes for predicting the biliary excretion of pharmaceutical and food-related compounds.J Toxicol Sci. 2023;48(5):251-261. doi: 10.2131/jts.48.251. J Toxicol Sci. 2023. PMID: 37121740
-
Bile canaliculi formation and biliary transport in 3D sandwich-cultured hepatocytes in dependence of the extracellular matrix composition.Arch Toxicol. 2016 Oct;90(10):2497-511. doi: 10.1007/s00204-016-1758-z. Epub 2016 Jun 21. Arch Toxicol. 2016. PMID: 27325308
-
Correlation of biliary excretion in sandwich-cultured rat hepatocytes and in vivo in rats.Drug Metab Dispos. 1999 Jun;27(6):637-44. Drug Metab Dispos. 1999. PMID: 10348791
-
Evaluation of Drug Biliary Excretion Using Sandwich-Cultured Human Hepatocytes.Eur J Drug Metab Pharmacokinet. 2019 Feb;44(1):13-30. doi: 10.1007/s13318-018-0502-x. Eur J Drug Metab Pharmacokinet. 2019. PMID: 30167999 Review.
-
[Frontline in Biliary Excretion of Drugs and Their Evaluation Systems].Yakugaku Zasshi. 2025;145(6):491-499. doi: 10.1248/yakushi.24-00165-1. Yakugaku Zasshi. 2025. PMID: 40451870 Review. Japanese.
References
-
- Tetsuka K, Ohbuchi M, Tabata K. Recent Progress in Hepatocyte Culture Models and Their Application to the Assessment of Drug Metabolism, Transport, and Toxicity in Drug Discovery: The Value of Tissue Engineering for the Successful Development of a Microphysiological System. J. Pharm. Sci. 2017;106:2302–2311. - PubMed
-
- Ohkura T, et al. Evaluation of human hepatocytes cultured by three-dimensional spheroid systems for drug metabolism. Drug Metabol. Pharmacokinet. 2014;29:373–378. - PubMed
-
- Arakawa H, et al. Kinetic analysis of sequential metabolism of triazolam and its extrapolation to humans using an entero-hepatic two-organ microphysiological system. Lab. Chip. 2020;20:537–547. - PubMed
-
- Scott DE, Bayly AR, Abell C, Skidmore J. Small molecules, big targets: drug discovery faces the protein-protein interaction challenge. Nat. Rev. Drug Discov. 2016;15:533–550. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

