Development of a workflow for the selection, identification and optimization of lactic acid bacteria with high γ-aminobutyric acid production
- PMID: 37608211
- PMCID: PMC10444875
- DOI: 10.1038/s41598-023-40808-z
Development of a workflow for the selection, identification and optimization of lactic acid bacteria with high γ-aminobutyric acid production
Abstract
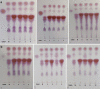
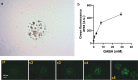
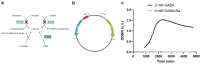
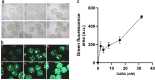
Lactic acid bacteria produce γ-aminobutyric acid (GABA) as an acid stress response. GABA is a neurotransmitter that may improve sleep and resilience to mental stress. This study focused on the selection, identification and optimization of a bacterial strain with high GABA production, for development as a probiotic supplement. The scientific literature and an industry database were searched for probiotics and potential GABA producers. In silico screening was conducted to identify genes involved in GABA production. Subsequently, 17 candidates were screened for in vitro GABA production using thin layer chromatography, which identified three candidate probiotic strains Levilactobacillus brevis DSM 20054, Lactococcus lactis DS75843and Bifidobacterium adolescentis DSM 24849 as producing GABA. Two biosensors capable of detecting GABA were developed: 1. a transcription factor-based biosensor characterized by the interaction with the transcriptional regulator GabR was developed in Corynebacterium glutamicum; and 2. a growth factor-based biosensor was built in Escherichia coli, which used auxotrophic complementation by expressing 4-aminobutyrate transaminase (GABA-T) that transfers the GABA amino group to pyruvate, hereby forming alanine. Consequently, the feasibility of developing a workflow based on co-culture with producer strains and a biosensor was tested. The three GABA producers were identified and the biosensors were encapsulated in nanoliter reactors (NLRs) as alginate beads in defined gut-like conditions. The E. coli growth factor-based biosensor was able to detect changes in GABA concentrations in liquid culture and under gut-like conditions. L. brevis and L. lactis were successfully encapsulated in the NLRs and showed growth under miniaturized intestinal conditions.
© 2023. Springer Nature Limited.
Conflict of interest statement
A.R., W.S., A.M., T.N.M. & G.S. are employees of dsm-firmenich. G.D.B, V.D and L.V. have no competing interests. J.K.B. is a consultant for dsm-firmenich/DSM Nutritional Products.
Figures
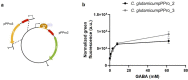

Similar articles
-
Gamma aminobutyric acid production by commercially available probiotic strains.J Appl Microbiol. 2023 Feb 16;134(2):lxac066. doi: 10.1093/jambio/lxac066. J Appl Microbiol. 2023. PMID: 36724279
-
Genome analysis and optimization of γ-aminobutyric acid (GABA) production by lactic acid bacteria from plant materials.J Gen Appl Microbiol. 2021 Oct 22;67(4):150-161. doi: 10.2323/jgam.2020.10.002. Epub 2021 Jun 5. J Gen Appl Microbiol. 2021. PMID: 34092710
-
Screening of lactic acid bacteria strains isolated from Iranian traditional dairy products for GABA production and optimization by response surface methodology.Sci Rep. 2023 Jan 9;13(1):440. doi: 10.1038/s41598-023-27658-5. Sci Rep. 2023. PMID: 36624130 Free PMC article.
-
Microbial chassis design and engineering for production of gamma-aminobutyric acid.World J Microbiol Biotechnol. 2024 Apr 12;40(5):159. doi: 10.1007/s11274-024-03951-x. World J Microbiol Biotechnol. 2024. PMID: 38607454 Review.
-
Production of Gamma-Aminobutyric Acid from Lactic Acid Bacteria: A Systematic Review.Int J Mol Sci. 2020 Feb 3;21(3):995. doi: 10.3390/ijms21030995. Int J Mol Sci. 2020. PMID: 32028587 Free PMC article.
Cited by
-
From bugs to brain: unravelling the GABA signalling networks in the brain-gut-microbiome axis.Brain. 2025 May 13;148(5):1479-1506. doi: 10.1093/brain/awae413. Brain. 2025. PMID: 39716883 Free PMC article. Review.
-
Biosynthesis of Gamma-Aminobutyric Acid (GABA) by Lactiplantibacillus plantarum in Fermented Food Production.Curr Issues Mol Biol. 2023 Dec 26;46(1):200-220. doi: 10.3390/cimb46010015. Curr Issues Mol Biol. 2023. PMID: 38248317 Free PMC article. Review.
-
HMOs Impact the Gut Microbiome of Children and Adults Starting from Low Predicted Daily Doses.Metabolites. 2024 Apr 20;14(4):239. doi: 10.3390/metabo14040239. Metabolites. 2024. PMID: 38668367 Free PMC article.
-
Are Aptamer-Based Biosensors the Future of the Detection of the Human Gut Microbiome?-A Systematic Review and Meta-Analysis.Biosensors (Basel). 2024 Sep 2;14(9):423. doi: 10.3390/bios14090423. Biosensors (Basel). 2024. PMID: 39329798 Free PMC article.
-
Investigation of different cold adaptation abilities in Salmonella enterica serotype Typhimurium strains using extracellular metabolomic approach.Int Microbiol. 2025 Mar;28(3):447-460. doi: 10.1007/s10123-024-00556-0. Epub 2024 Jul 8. Int Microbiol. 2025. PMID: 38977514
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

