This is a preprint.
PHOSPHORYLATION-DEPENDENT ASSOCIATION OF WRN WITH RPA IS REQUIRED FOR RECOVERY OF REPLICATION FORKS STALLED AT SECONDARY DNA STRUCTURES
- PMID: 37609214
- PMCID: PMC10441285
- DOI: 10.1101/2023.08.08.552428
PHOSPHORYLATION-DEPENDENT ASSOCIATION OF WRN WITH RPA IS REQUIRED FOR RECOVERY OF REPLICATION FORKS STALLED AT SECONDARY DNA STRUCTURES
Update in
-
Phosphorylation-dependent WRN-RPA interaction promotes recovery of stalled forks at secondary DNA structure.Nat Commun. 2025 Jan 27;16(1):997. doi: 10.1038/s41467-025-55958-z. Nat Commun. 2025. PMID: 39870632 Free PMC article.
Abstract
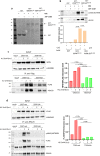
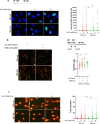
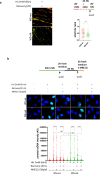
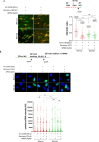
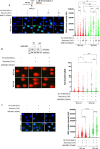
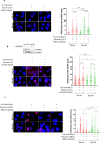
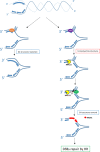
The WRN protein mutated in the hereditary premature aging disorder Werner syndrome plays a vital role in handling, processing, and restoring perturbed replication forks. One of its most abundant partners, Replication Protein A (RPA), has been shown to robustly enhance WRN helicase activity in specific cases when tested in vitro. However, the significance of RPA-binding to WRN at replication forks in vivo has remained largely unexplored. In this study, we have identified several conserved phosphorylation sites in the acidic domain of WRN that are targeted by Casein Kinase 2 (CK2). Surprisingly, these phosphorylation sites are essential for the interaction between WRN and RPA, both in vitro and in human cells. By characterizing a CK2-unphosphorylatable WRN mutant that lacks the ability to bind RPA, we have determined that the WRN-RPA complex plays a critical role in fork recovery after replication stress whereas the WRN-RPA interaction is not necessary for the processing of replication forks or preventing DNA damage when forks stall or collapse. When WRN fails to bind RPA, fork recovery is impaired, leading to the accumulation of single-stranded DNA gaps in the parental strands, which are further enlarged by the structure-specific nuclease MRE11. Notably, RPA-binding by WRN and its helicase activity are crucial for countering the persistence of G4 structures after fork stalling. Therefore, our findings reveal for the first time a novel role for the WRN-RPA interaction to facilitate fork restart, thereby minimizing G4 accumulation at single-stranded DNA gaps and suppressing accumulation of unreplicated regions that may lead to MUS81-dependent double-strand breaks requiring efficient repair by RAD51 to prevent excessive DNA damage.
Keywords: Genome Instability; RecQ helicases; Replication fork arrest; Werner syndrome.
Conflict of interest statement
CONFLICT OF INTEREST The authors declare to do not have any conflict of interest.
Figures






References
-
- Franchitto A., Pichierri P., Understanding the molecular basis of common fragile sites instability: Role of the proteins involved in the recovery of stalled replication forks. Cell Cycle. 10, 4039–4046 (2011). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
