This is a preprint.
Glial-derived mitochondrial signals impact neuronal proteostasis and aging
- PMID: 37609253
- PMCID: PMC10441375
- DOI: 10.1101/2023.07.20.549924
Glial-derived mitochondrial signals impact neuronal proteostasis and aging
Update in
-
Glial-derived mitochondrial signals affect neuronal proteostasis and aging.Sci Adv. 2023 Oct 13;9(41):eadi1411. doi: 10.1126/sciadv.adi1411. Epub 2023 Oct 13. Sci Adv. 2023. PMID: 37831769 Free PMC article.
Abstract
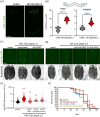
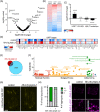
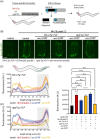
The nervous system plays a critical role in maintaining whole-organism homeostasis; neurons experiencing mitochondrial stress can coordinate the induction of protective cellular pathways, such as the mitochondrial unfolded protein response (UPRMT), between tissues. However, these studies largely ignored non-neuronal cells of the nervous system. Here, we found that UPRMT activation in four, astrocyte-like glial cells in the nematode, C. elegans, can promote protein homeostasis by alleviating protein aggregation in neurons. Surprisingly, we find that glial cells utilize small clear vesicles (SCVs) to signal to neurons, which then relay the signal to the periphery using dense-core vesicles (DCVs). This work underlines the importance of glia in establishing and regulating protein homeostasis within the nervous system, which can then impact neuron-mediated effects in organismal homeostasis and longevity.
Keywords: C. elegans; aging; glia; mitochondria; neurodegeneration; protein homeostasis.
Conflict of interest statement
Competing interests: All authors of the manuscript declare that they have no competing interests.
Figures



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
