GADMA2: more efficient and flexible demographic inference from genetic data
- PMID: 37609916
- PMCID: PMC10445054
- DOI: 10.1093/gigascience/giad059
GADMA2: more efficient and flexible demographic inference from genetic data
Abstract
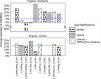
Background: Inference of complex demographic histories is a source of information about events that happened in the past of studied populations. Existing methods for demographic inference typically require input from the researcher in the form of a parameterized model. With an increased variety of methods and tools, each with its own interface, the model specification becomes tedious and error-prone. Moreover, optimization algorithms used to find model parameters sometimes turn out to be inefficient, for instance, by being not properly tuned or highly dependent on a user-provided initialization. The open-source software GADMA addresses these problems, providing automatic demographic inference. It proposes a common interface for several likelihood engines and provides global parameters optimization based on a genetic algorithm.
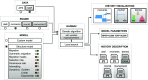
Results: Here, we introduce the new GADMA2 software and provide a detailed description of the added and expanded features. It has a renovated core code base, new likelihood engines, an updated optimization algorithm, and a flexible setup for automatic model construction. We provide a full overview of GADMA2 enhancements, compare the performance of supported likelihood engines on simulated data, and demonstrate an example of GADMA2 usage on 2 empirical datasets.
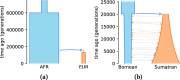
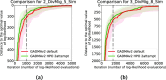
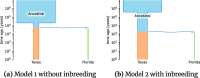
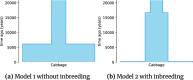
Conclusions: We demonstrate the better performance of a genetic algorithm in GADMA2 by comparing it to the initial version and other existing optimization approaches. Our experiments on simulated data indicate that GADMA2's likelihood engines are able to provide accurate estimations of demographic parameters even for misspecified models. We improve model parameters for 2 empirical datasets of inbred species.
Keywords: demographic inference; genetic algorithm; hyperparameter optimization; population genetics.
© The Author(s) 2023. Published by Oxford University Press GigaScience.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

