Predicting ion mobility collision cross sections using projection approximation with ROSIE-PARCS webserver
- PMID: 37609950
- PMCID: PMC10516336
- DOI: 10.1093/bib/bbad308
Predicting ion mobility collision cross sections using projection approximation with ROSIE-PARCS webserver
Abstract
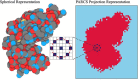
Ion mobility coupled to mass spectrometry informs on the shape and size of protein structures in the form of a collision cross section (CCSIM). Although there are several computational methods for predicting CCSIM based on protein structures, including our previously developed projection approximation using rough circular shapes (PARCS), the process usually requires prior experience with the command-line interface. To overcome this challenge, here we present a web application on the Rosetta Online Server that Includes Everyone (ROSIE) webserver to predict CCSIM from protein structure using projection approximation with PARCS. In this web interface, the user is only required to provide one or more PDB files as input. Results from our case studies suggest that CCSIM predictions (with ROSIE-PARCS) are highly accurate with an average error of 6.12%. Furthermore, the absolute difference between CCSIM and CCSPARCS can help in distinguishing accurate from inaccurate AlphaFold2 protein structure predictions. ROSIE-PARCS is designed with a user-friendly interface, is available publicly and is free to use. The ROSIE-PARCS web interface is supported by all major web browsers and can be accessed via this link (https://rosie.graylab.jhu.edu).
Keywords: AlphaFold2; collision cross section; ion mobility; mass spectrometry; protein structure prediction; webserver.
© The Author(s) 2023. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures




References
-
- Wyttenbach T, Bowers MT. Structural stability from solution to the gas phase: native solution structure of ubiquitin survives analysis in a solvent-free ion mobility-mass spectrometry environment. J Phys Chem B 2011;115:12266–75. - PubMed
-
- Ruotolo BT, Robinson CV. Aspects of native proteins are retained in vacuum. Curr Opin Chem Biol 2006;10:402–8. - PubMed
-
- Bleiholder C, Liu FC. Structure relaxation approximation (SRA) for elucidation of protein structures from ion mobility measurements. J Phys Chem B 2019;123:2756–69. - PubMed
-
- Leney AC, Heck AJR. Native mass spectrometry: what is in the name? J Am Soc Mass Spectrom 2017;28:5–13. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

