Staphylococcus epidermidis activates keratinocyte cytokine expression and promotes skin inflammation through the production of phenol-soluble modulins
- PMID: 37610872
- PMCID: PMC10586132
- DOI: 10.1016/j.celrep.2023.113024
Staphylococcus epidermidis activates keratinocyte cytokine expression and promotes skin inflammation through the production of phenol-soluble modulins
Abstract
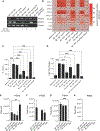
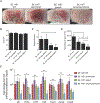
Staphylococcus epidermidis is a common microbe on human skin and has beneficial functions in the skin microbiome. However, under conditions of allergic inflammation, the abundance of S. epidermidis increases, establishing potential danger to the epidermis. To understand how this commensal may injure the host, we investigate phenol-soluble modulin (PSM) peptides produced by S. epidermidis that are similar to peptides produced by Staphylococcus aureus. Synthetic S. epidermidis PSMs induce expression of host defense genes and are cytotoxic to human keratinocytes. Deletion mutants of S. epidermidis lacking these gene products support these observations and further show that PSMs require the action of the EcpA bacterial protease to induce inflammation when applied on mouse skin with an intact stratum corneum. The expression of PSMδ from S. epidermidis is also found to correlate with disease severity in patients with atopic dermatitis. These observations show how S. epidermidis PSMs can promote skin inflammation.
Keywords: CP: Immunology; CP: Microbiology; Staphylococcus; atopic dermatitis; inflammation; microbiome; phenol-soluble modulins; skin.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests R.L.G. is a co-founder, scientific advisor, and consultant and has equity in MatriSys Biosciences.
Figures



References
-
- Cogen AL, Yamasaki K, Sanchez KM, Dorschner RA, Lai Y, MacLeod DT, Torpey JW, Otto M, Nizet V, Kim JE, and Gallo RL (2010). Selective antimicrobial action is provided by phenol-soluble modulins derived from Staphylococcus epidermidis, a normal resident of the skin. J. Invest. Dermatol. 130, 192–200. 10.1038/jid.2009.243. - DOI - PMC - PubMed
-
- Williams MR, Costa SK, Zaramela LS, Khalil S, Todd DA, Winter HL, Sanford JA, O’Neill AM, Liggins MC, Nakatsuji T, et al. (2019). Quorum sensing between bacterial species on the skin protects against epidermal injury in atopic dermatitis. Sci. Transl. Med. 11, eaat8329. 10.1126/scitranslmed.aat8329. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

