A MYCN-independent mechanism mediating secretome reprogramming and metastasis in MYCN-amplified neuroblastoma
- PMID: 37611092
- PMCID: PMC10446492
- DOI: 10.1126/sciadv.adg6693
A MYCN-independent mechanism mediating secretome reprogramming and metastasis in MYCN-amplified neuroblastoma
Abstract
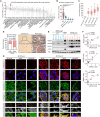
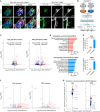
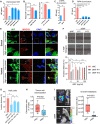
MYCN amplification (MNA) is a defining feature of high-risk neuroblastoma (NB) and predicts poor prognosis. However, whether genes within or in close proximity to the MYCN amplicon also contribute to MNA+ NB remains poorly understood. Here, we identify that GREB1, a transcription factor encoding gene neighboring the MYCN locus, is frequently coexpressed with MYCN and promotes cell survival in MNA+ NB. GREB1 controls gene expression independently of MYCN, among which we uncover myosin 1B (MYO1B) as being highly expressed in MNA+ NB and, using a chick chorioallantoic membrane (CAM) model, as a crucial regulator of invasion and metastasis. Global secretome and proteome profiling further delineates MYO1B in regulating secretome reprogramming in MNA+ NB cells, and the cytokine MIF as an important pro-invasive and pro-metastatic mediator of MYO1B activity. Together, we have identified a putative GREB1-MYO1B-MIF axis as an unconventional mechanism promoting aggressive behavior in MNA+ NB and independently of MYCN.
Figures



References
-
- K. K. Matthay, J. M. Maris, G. Schleiermacher, A. Nakagawara, C. L. Mackall, L. Diller, W. A. Weiss, Neuroblastoma. Nat. Rev. Dis. Primers 2, 16078 (2016). - PubMed
-
- G. M. Brodeur, R. C. Seeger, M. Schwab, H. E. Varmus, J. M. Bishop, Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science 224, 1121–1124 (1984). - PubMed
-
- S. L. Cohn, A. D. J. Pearson, W. B. London, T. Monclair, P. F. Ambros, G. M. Brodeur, A. Faldum, B. Hero, T. Iehara, D. Machin, V. Mosseri, T. Simon, A. Garaventa, V. Castel, K. K. Matthay; INRG Task Force , The International Neuroblastoma Risk Group (INRG) classification system: An INRG Task Force report. J. Clin. Oncol. 27, 289–297 (2009). - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

