hMENA isoforms regulate cancer intrinsic type I IFN signaling and extrinsic mechanisms of resistance to immune checkpoint blockade in NSCLC
- PMID: 37612043
- PMCID: PMC10450042
- DOI: 10.1136/jitc-2023-006913
hMENA isoforms regulate cancer intrinsic type I IFN signaling and extrinsic mechanisms of resistance to immune checkpoint blockade in NSCLC
Abstract
Background: Understanding how cancer signaling pathways promote an immunosuppressive program which sustains acquired or primary resistance to immune checkpoint blockade (ICB) is a crucial step in improving immunotherapy efficacy. Among the pathways that can affect ICB response is the interferon (IFN) pathway that may be both detrimental and beneficial. The immune sensor retinoic acid-inducible gene I (RIG-I) induces IFN activation and secretion and is activated by actin cytoskeleton disturbance. The actin cytoskeleton regulatory protein hMENA, along with its isoforms, is a key signaling hub in different solid tumors, and recently its role as a regulator of transcription of genes encoding immunomodulatory secretory proteins has been proposed. When hMENA is expressed in tumor cells with low levels of the epithelial specific hMENA11a isoform, identifies non-small cell lung cancer (NSCLC) patients with poor prognosis. Aim was to identify cancer intrinsic and extrinsic pathways regulated by hMENA11a downregulation as determinants of ICB response in NSCLC. Here, we present a potential novel mechanism of ICB resistance driven by hMENA11a downregulation.
Methods: Effects of hMENA11a downregulation were tested by RNA-Seq, ATAC-Seq, flow cytometry and biochemical assays. ICB-treated patient tumor tissues were profiled by Nanostring IO 360 Panel enriched with hMENA custom probes. OAK and POPLAR datasets were used to validate our discovery cohort.
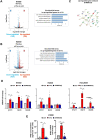
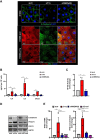
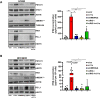
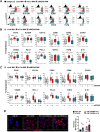
Results: Transcriptomic and biochemical analyses demonstrated that the depletion of hMENA11a induces IFN pathway activation, the production of different inflammatory mediators including IFNβ via RIG-I, sustains the increase of tumor PD-L1 levels and activates a paracrine loop between tumor cells and a unique macrophage subset favoring an epithelial-mesenchymal transition (EMT). Notably, when we translated our results in a clinical setting of NSCLC ICB-treated patients, transcriptomic analysis revealed that low expression of hMENA11a, high expression of IFN target genes and high macrophage score identify patients resistant to ICB therapy.
Conclusions: Collectively, these data establish a new function for the actin cytoskeleton regulator hMENA11a in modulating cancer cell intrinsic type I IFN signaling and extrinsic mechanisms that promote protumoral macrophages and favor EMT. These data highlight the role of actin cytoskeleton disturbance in activating immune suppressive pathways that may be involved in resistance to ICB in NSCLC.
Keywords: biomarkers, tumor; cytokines; immune checkpoint inhibitors; macrophages; non-small cell lung cancer.
© Author(s) (or their employer(s)) 2023. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
