RNA modification: mechanisms and therapeutic targets
- PMID: 37612540
- PMCID: PMC10447785
- DOI: 10.1186/s43556-023-00139-x
RNA modification: mechanisms and therapeutic targets
Abstract
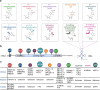
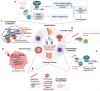
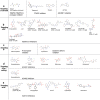
RNA modifications are dynamic and reversible chemical modifications on substrate RNA that are regulated by specific modifying enzymes. They play important roles in the regulation of many biological processes in various diseases, such as the development of cancer and other diseases. With the help of advanced sequencing technologies, the role of RNA modifications has caught increasing attention in human diseases in scientific research. In this review, we briefly summarized the basic mechanisms of several common RNA modifications, including m6A, m5C, m1A, m7G, Ψ, A-to-I editing and ac4C. Importantly, we discussed their potential functions in human diseases, including cancer, neurological disorders, cardiovascular diseases, metabolic diseases, genetic and developmental diseases, as well as immune disorders. Through the "writing-erasing-reading" mechanisms, RNA modifications regulate the stability, translation, and localization of pivotal disease-related mRNAs to manipulate disease development. Moreover, we also highlighted in this review all currently available RNA-modifier-targeting small molecular inhibitors or activators, most of which are designed against m6A-related enzymes, such as METTL3, FTO and ALKBH5. This review provides clues for potential clinical therapy as well as future study directions in the RNA modification field. More in-depth studies on RNA modifications, their roles in human diseases and further development of their inhibitors or activators are needed for a thorough understanding of epitranscriptomics as well as diagnosis, treatment, and prognosis of human diseases.
Keywords: Cancer; Cardiovascular diseases; Inhibitors; Metabolic diseases; Neurological disorders; RNA modification.
© 2023. Sichuan International Medical Exchange & Promotion Association.
Conflict of interest statement
The authors declare no conflicts of interest with the contents of this article.
Figures




References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
