Alterations of lipid-mediated mitophagy result in aging-dependent sensorimotor defects
- PMID: 37614052
- PMCID: PMC10577547
- DOI: 10.1111/acel.13954
Alterations of lipid-mediated mitophagy result in aging-dependent sensorimotor defects
Abstract
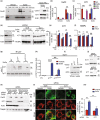
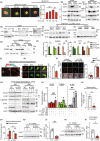
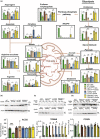
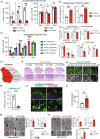
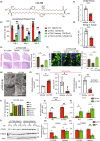
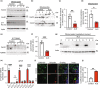
The metabolic consequences of mitophagy alterations due to age-related stress in healthy aging brains versus neurodegeneration remain unknown. Here, we demonstrate that ceramide synthase 1 (CerS1) is transported to the outer mitochondrial membrane by the p17/PERMIT transporter that recognizes mislocalized mitochondrial ribosomes (mitoribosomes) via 39-FLRN-42 residues, inducing ceramide-mediated mitophagy. P17/PERMIT-CerS1-mediated mitophagy attenuated the argininosuccinate/fumarate/malate axis and induced d-glucose and fructose accumulation in neurons in culture and brain tissues (primarily in the cerebellum) of wild-type mice in vivo. These metabolic changes in response to sodium-selenite were nullified in the cerebellum of CerS1to/to (catalytically inactive for C18-ceramide production CerS1 mutant), PARKIN-/- or p17/PERMIT-/- mice that have dysfunctional mitophagy. Whereas sodium selenite induced mitophagy in the cerebellum and improved motor-neuron deficits in aged wild-type mice, exogenous fumarate or malate prevented mitophagy. Attenuating ceramide-mediated mitophagy enhanced damaged mitochondria accumulation and age-dependent sensorimotor abnormalities in p17/PERMIT-/- mice. Reinstituting mitophagy using a ceramide analog drug with selenium conjugate, LCL768, restored mitophagy and reduced malate/fumarate metabolism, improving sensorimotor deficits in old p17/PERMIT-/- mice. Thus, these data describe the metabolic consequences of alterations to p17/PERMIT/ceramide-mediated mitophagy associated with the loss of mitochondrial quality control in neurons and provide therapeutic options to overcome age-dependent sensorimotor deficits and related disorders like amyotrophic lateral sclerosis (ALS).
Keywords: CerS1; Drp1; aging; ceramide; mitochondrial metabolism; mitophagy; neurodegeneration; sensorimotor defects.
© 2023 The Authors. Aging Cell published by Anatomical Society and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Bede, P. , Chipika, R. H. , Christidi, F. , Hengeveld, J. C. , Karavasilis, E. , Argyropoulos, G. D. , Lope, J. , Li Hi Shing, S. , Velonakis, G. , Dupuis, L. , Doherty, M. A. , Vajda, A. , McLaughlin, R. , & Hardiman, O. (2021). Genotype‐associated cerebellar profiles in ALS: Focal cerebellar pathology and cerebro‐cerebellar connectivity alterations. Journal of Neurology, Neurosurgery and Psychiatry, 92(11), 1197–1205. 10.1136/jnnp-2021-326854 - DOI - PMC - PubMed
-
- Cheng, C. T. , Qi, Y. , Wang, Y. C. , Chi, K. K. , Chung, Y. , Ouyang, C. , Ann, D. K. , Oh, M. E. , Sheng, X. , Tang, Y. , Liu, Y. R. , Lin, H. H. , Kuo, C. Y. , Schones, D. , Vidal, C. M. , Chu, J. C. , Wang, H. J. , Chen, Y. H. , Miller, K. M. , … Ann, D. K. (2018). Arginine starvation kills tumor cells through aspartate exhaustion and mitochondrial dysfunction. Communications Biology, 1, 178. 10.1038/s42003-018-0178-4 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

