METTL14 inhibits malignant progression of oral squamous cell carcinoma by targeting the autophagy-related gene RB1CC1 in an m6A-IGF2BP2-dependent manner
- PMID: 37615536
- PMCID: PMC10500204
- DOI: 10.1042/CS20230219
METTL14 inhibits malignant progression of oral squamous cell carcinoma by targeting the autophagy-related gene RB1CC1 in an m6A-IGF2BP2-dependent manner
Abstract
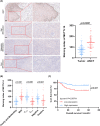
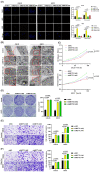
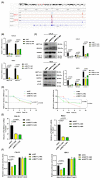
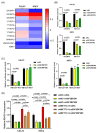
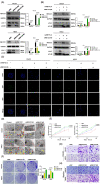
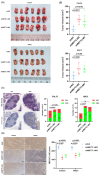
N6-methyladenosine (m6A) plays crucial roles in tumorigenesis and autophagy. However, the underlying mechanisms mediated by m6A and autophagy in the malignant progression of oral squamous cell carcinoma (OSCC) remain unclear. In the present study, we revealed that down-regulated expression of METTL14 was correlated with advanced clinicopathological characteristics and poor prognosis in OSCC. METTL14 knockdown significantly inhibited autophagy and facilitated malignant progression in vitro, and promoted tumor growth and metastasis in vivo. A cell model of rapamycin-induced autophagy was established to identify RB1CC1 as a potential target gene involved in m6A-regulated autophagy in OSCC, through RNA sequencing and methylated RNA immunoprecipitation sequencing (meRIP-seq) analysis. Mechanistically, we confirmed that METTL14 posttranscriptionally enhanced RB1CC1 expression in an m6A-IGF2BP2-dependent manner, thereby affecting autophagy and progression in OSCC, through methylated RNA immunoprecipitation qRT-PCR (meRIP-qPCR), RNA stability assays, mutagenesis assays and dual-luciferase reporter. Collectively, our findings demonstrated that METTL14 serves as an OSCC suppressor by regulating the autophagy-related gene RB1CC1 through m6A modification, which may provide a new insight for the diagnosis and therapy of OSCC.
Keywords: autophagy; epigenetics; methyltransferase-like 14; oncogenesis; oral squamous cell carcinoma.
© 2023 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures
Similar articles
-
Mechanism of METTL14 and m6A modification of lncRNA MALAT1 in the proliferation of oral squamous cell carcinoma cells.Oral Dis. 2023 Jul;29(5):2012-2026. doi: 10.1111/odi.14220. Epub 2022 May 17. Oral Dis. 2023. PMID: 35467063
-
METTL14 Promotes Oral Squamous Cell Carcinoma Progression by Regulating the mRNA and m6A Levels of CALD1.J Environ Pathol Toxicol Oncol. 2023;42(3):71-81. doi: 10.1615/JEnvironPatholToxicolOncol.2022045134. J Environ Pathol Toxicol Oncol. 2023. PMID: 37017680
-
RNA N6-methyladenosine reader IGF2BP2 promotes lymphatic metastasis and epithelial-mesenchymal transition of head and neck squamous carcinoma cells via stabilizing slug mRNA in an m6A-dependent manner.J Exp Clin Cancer Res. 2022 Jan 3;41(1):6. doi: 10.1186/s13046-021-02212-1. J Exp Clin Cancer Res. 2022. PMID: 34980207 Free PMC article.
-
Insights into roles of METTL14 in tumors.Cell Prolif. 2022 Jan;55(1):e13168. doi: 10.1111/cpr.13168. Epub 2021 Dec 13. Cell Prolif. 2022. PMID: 34904301 Free PMC article. Review.
-
[Research progress on the role and mechanism of IGF2BPs family in head and neck squamous carcinoma].Lin Chuang Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2024 Dec;38(12):1195-1202. doi: 10.13201/j.issn.2096-7993.2024.12.021. Lin Chuang Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2024. PMID: 39605275 Free PMC article. Review. Chinese.
Cited by
-
Decoding the epitranscriptome: a new frontier for cancer therapy and drug resistance.Cell Commun Signal. 2024 Oct 21;22(1):513. doi: 10.1186/s12964-024-01854-w. Cell Commun Signal. 2024. PMID: 39434167 Free PMC article. Review.
-
Mettl3-Mediated N6-Methyladenosine Modification Mitigates Ganglion Cell Loss and Retinal Dysfunction in Retinal Ischemia-Reperfusion Injury by Inhibiting FoxO1-Mediated Autophagy.Invest Ophthalmol Vis Sci. 2025 Feb 3;66(2):58. doi: 10.1167/iovs.66.2.58. Invest Ophthalmol Vis Sci. 2025. PMID: 39982709 Free PMC article.
-
The role of N(6)-methyladenosine (m6a) modification in cancer: recent advances and future directions.EXCLI J. 2025 Jan 15;24:113-150. doi: 10.17179/excli2024-7935. eCollection 2025. EXCLI J. 2025. PMID: 39967906 Free PMC article. Review.
-
SMAD specific E3 ubiquitin protein ligase 1 accelerates diabetic macular edema progression by WNT inhibitory factor 1.World J Diabetes. 2025 Mar 15;16(3):101328. doi: 10.4239/wjd.v16.i3.101328. World J Diabetes. 2025. PMID: 40093288 Free PMC article.
-
Targeting autophagy to enhance chemotherapy and immunotherapy in oral cancer.Front Immunol. 2025 Jan 7;15:1535649. doi: 10.3389/fimmu.2024.1535649. eCollection 2024. Front Immunol. 2025. PMID: 39840028 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

