Trained immunity of alveolar macrophages enhances injury resolution via KLF4-MERTK-mediated efferocytosis
- PMID: 37615937
- PMCID: PMC10450795
- DOI: 10.1084/jem.20221388
Trained immunity of alveolar macrophages enhances injury resolution via KLF4-MERTK-mediated efferocytosis
Abstract
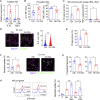
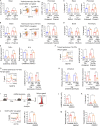
Recent studies suggest that training of innate immune cells such as tissue-resident macrophages by repeated noxious stimuli can heighten host defense responses. However, it remains unclear whether trained immunity of tissue-resident macrophages also enhances injury resolution to counterbalance the heightened inflammatory responses. Here, we studied lung-resident alveolar macrophages (AMs) prechallenged with either the bacterial endotoxin or with Pseudomonas aeruginosa and observed that these trained AMs showed greater resilience to pathogen-induced cell death. Transcriptomic analysis and functional assays showed greater capacity of trained AMs for efferocytosis of cellular debris and injury resolution. Single-cell high-dimensional mass cytometry analysis and lineage tracing demonstrated that training induces an expansion of a MERTKhiMarcohiCD163+F4/80low lung-resident AM subset with a proresolving phenotype. Reprogrammed AMs upregulated expression of the efferocytosis receptor MERTK mediated by the transcription factor KLF4. Adoptive transfer of these trained AMs restricted inflammatory lung injury in recipient mice exposed to lethal P. aeruginosa. Thus, our study has identified a subset of tissue-resident trained macrophages that prevent hyperinflammation and restore tissue homeostasis following repeated pathogen challenges.
© 2023 Chakraborty et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures












References
-
- Amir, el-A.D., Davis K.L., Tadmor M.D., Simonds E.F., Levine J.H., Bendall S.C., Shenfeld D.K., Krishnaswamy S., Nolan G.P., and Pe’er D.. 2013. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat. Biotechnol. 31:545–552. 10.1038/nbt.2594 - DOI - PMC - PubMed
-
- Ampomah, P.B., Cai B., Sukka S.R., Gerlach B.D., Yurdagul A. Jr, Wang X., Kuriakose G., Darville L.N.F., Sun Y., Sidoli S., et al. . 2022. Macrophages use apoptotic cell-derived methionine and DNMT3A during efferocytosis to promote tissue resolution. Nat. Metab. 4:444–457. 10.1038/s42255-022-00551-7 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

