Integrative analysis links autophagy to intrauterine adhesion and establishes autophagy-related circRNA-miRNA-mRNA regulatory network
- PMID: 37616056
- PMCID: PMC10497020
- DOI: 10.18632/aging.204969
Integrative analysis links autophagy to intrauterine adhesion and establishes autophagy-related circRNA-miRNA-mRNA regulatory network
Abstract
Background: Intrauterine adhesion (IUA) is a troublesome complication characterized with endometrial fibrosis after endometrial trauma. Increasing number of investigations focused on autophagy and non-coding RNA in the pathogenesis of uterine adhesion, but the underlying mechanism needs to be further studied.
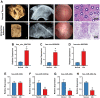
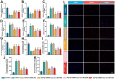
Methods: mRNA expression profile and miRNA expression profile were obtained from Gene Expression Omnibus database. The autophagy related genes were low. Venn diagram was used to set the intersection of autophagy genes and DEGs to obtain ARDEGs. Circbank was used to select hub autophagy-related circRNAs based on ARDEMs. Then, the differentially expressed autophagy-related genes, miRNAs and circRNAs were analyzed by functional enrichment analysis, and protein-protein interaction network analysis. Finally, the expression levels of hub circRNAs and hub miRNAs were validated through RT-PCR of clinical intrauterine adhesion samples. In vitro experiments were investigated to explore the effect of hub ARCs on cell autophagy, myofibroblast transformation and collagen deposition.
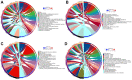
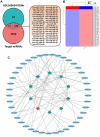
Results: 11 autophagy-related differentially expressed genes (ARDEGs) and 41 differentially expressed miRNA (ARDEMs) compared between normal tissues and IUA were identified. Subsequently, the autophagy-related miRNA-mRNA network was constructed and hub ARDEMs were selected. Furthermore, the autophagy-related circRNA-miRNA-mRNA network was established. According to the ranking of number of regulated ARDEMs, hsa-circ-0047959, hsa-circ-0032438, hsa-circ-0047301 were regarded as the hub ARCs. In comparison of normal endometrial tissue, all three hub ARCs were upregulated in IUA tissue. All hub ARDEMs were downregulated except has-miR-320c.
Conclusions: In the current study, we firstly constructed autophagy-related circRNA-miRNA-mRNA regulatory network and identified hub ARCs and ARDEMs had not been reported in IUA.
Keywords: autophagy; bioinformatics; circRNA-miRNA-mRNA; intrauterine adhesion; regulatory network.
Conflict of interest statement
Figures






Similar articles
-
Construction of the circRNA-miRNA-mRNA Regulatory Network of an Abdominal Aortic Aneurysm to Explore Its Potential Pathogenesis.Dis Markers. 2021 Nov 5;2021:9916881. doi: 10.1155/2021/9916881. eCollection 2021. Dis Markers. 2021. PMID: 34777635 Free PMC article.
-
Comprehensive Analysis of circRNA-miRNA-mRNA Regulatory Network and Novel Potential Biomarkers in Acute Myocardial Infarction.Front Cardiovasc Med. 2022 Jul 7;9:850991. doi: 10.3389/fcvm.2022.850991. eCollection 2022. Front Cardiovasc Med. 2022. PMID: 35872921 Free PMC article.
-
Construct a circRNA/miRNA/mRNA regulatory network to explore potential pathogenesis and therapy options of clear cell renal cell carcinoma.Sci Rep. 2020 Aug 12;10(1):13659. doi: 10.1038/s41598-020-70484-2. Sci Rep. 2020. PMID: 32788609 Free PMC article.
-
Construction and Bioinformatics Analysis of circRNA-miRNA-mRNA Network in Acute Myocardial Infarction.Front Genet. 2022 Mar 29;13:854993. doi: 10.3389/fgene.2022.854993. eCollection 2022. Front Genet. 2022. PMID: 35422846 Free PMC article.
-
Identification of potentially functional circular RNAs hsa_circ_0070934 and hsa_circ_0004315 as prognostic factors of hepatocellular carcinoma by integrated bioinformatics analysis.Sci Rep. 2022 Mar 23;12(1):4933. doi: 10.1038/s41598-022-08867-w. Sci Rep. 2022. PMID: 35322101 Free PMC article.
Cited by
-
Exploring NamiRNA networks and time-series gene expression in osteogenic differentiation of adipose-derived stem cells.Ann Med. 2025 Dec;57(1):2478323. doi: 10.1080/07853890.2025.2478323. Epub 2025 Mar 18. Ann Med. 2025. PMID: 40100054 Free PMC article.
-
ULK1 methylation promotes TGF-β1-induced endometrial fibrosis via the FOXP1/DNMT1 axis.Kaohsiung J Med Sci. 2025 Jan;41(1):e12915. doi: 10.1002/kjm2.12915. Epub 2024 Dec 4. Kaohsiung J Med Sci. 2025. PMID: 39629895 Free PMC article.
-
Mechanistic insights into intrauterine adhesions.Semin Immunopathol. 2024 Nov 29;47(1):3. doi: 10.1007/s00281-024-01030-9. Semin Immunopathol. 2024. PMID: 39613882 Review.
-
Crosstalk of ubiquitin system and non-coding RNA in fibrosis.Int J Biol Sci. 2024 Jul 8;20(10):3802-3822. doi: 10.7150/ijbs.93644. eCollection 2024. Int J Biol Sci. 2024. PMID: 39113708 Free PMC article. Review.
-
Gene Therapy for Inflammatory Cascade in Intrauterine Injury with Engineered Extracellular Vesicles Hybrid Snail Mucus-enhanced Adhesive Hydrogels.Adv Sci (Weinh). 2025 Jan;12(1):e2410769. doi: 10.1002/advs.202410769. Epub 2024 Oct 25. Adv Sci (Weinh). 2025. PMID: 39454114 Free PMC article.
References
-
- Wang S, Lu M, Cao Y, Tao Z, Sun Z, Liu X, Liu J, Li S. Degradative polylactide nanofibers promote M2 macrophage polarization via STAT6 pathway in peritendinous adhesion. Composites Part B: Engineering. 2023; 253:110520. 10.1016/j.compositesb.2023.110520 - DOI
-
- Zhang X, Liu W, Zhou Y, Qiu J, Sun Y, Li M, Ding Y, Xi Q. Comparison of therapeutic efficacy of three methods to prevent re-adhesion after hysteroscopic intrauterine adhesion separation: a parallel, randomized and single-center trial. Ann Palliat Med. 2021; 10:6804–23. 10.21037/apm-21-1296 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

