Cdc73 protects Notch-induced T-cell leukemia cells from DNA damage and mitochondrial stress
- PMID: 37616559
- PMCID: PMC10733839
- DOI: 10.1182/blood.2023020144
Cdc73 protects Notch-induced T-cell leukemia cells from DNA damage and mitochondrial stress
Erratum in
-
Melnick AF, Mullin C, Lin K, et al. Cdc73 protects Notch-induced T-cell leukemia cells from DNA damage and mitochondrial stress. Blood. 2023;142(25):2159-2174.Blood. 2024 Jul 18;144(3):342. doi: 10.1182/blood.2024025365. Blood. 2024. PMID: 39023871 Free PMC article. No abstract available.
Abstract
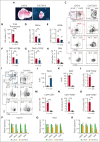
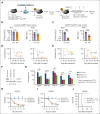
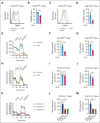
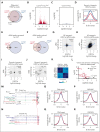
Activated Notch signaling is highly prevalent in T-cell acute lymphoblastic leukemia (T-ALL), but pan-Notch inhibitors showed excessive toxicity in clinical trials. To find alternative ways to target Notch signals, we investigated cell division cycle 73 (Cdc73), which is a Notch cofactor and key component of the RNA polymerase-associated transcriptional machinery, an emerging target in T-ALL. Although we confirmed previous work that CDC73 interacts with NOTCH1, we also found that the interaction in T-ALL was context-dependent and facilitated by the transcription factor ETS1. Using mouse models, we showed that Cdc73 is important for Notch-induced T-cell development and T-ALL maintenance. Mechanistically, chromatin and nascent gene expression profiling showed that Cdc73 intersects with Ets1 and Notch at chromatin within enhancers to activate expression of known T-ALL oncogenes through its enhancer functions. Cdc73 also intersects with these factors within promoters to activate transcription of genes that are important for DNA repair and oxidative phosphorylation through its gene body functions. Consistently, Cdc73 deletion induced DNA damage and apoptosis and impaired mitochondrial function. The CDC73-induced DNA repair expression program co-opted by NOTCH1 is more highly expressed in T-ALL than in any other cancer. These data suggest that Cdc73 might induce a gene expression program that was eventually intersected and hijacked by oncogenic Notch to augment proliferation and mitigate the genotoxic and metabolic stresses of elevated Notch signaling. Our report supports studying factors such as CDC73 that intersect with Notch to derive a basic scientific understanding on how to combat Notch-dependent cancers without directly targeting the Notch complex.
© 2023 by The American Society of Hematology.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures




Update of
-
Cdc73 protects Notch-induced T-cell leukemia cells from DNA damage and mitochondrial stress.bioRxiv [Preprint]. 2023 Feb 4:2023.01.22.525059. doi: 10.1101/2023.01.22.525059. bioRxiv. 2023. Update in: Blood. 2023 Dec 21;142(25):2159-2174. doi: 10.1182/blood.2023020144. PMID: 36711472 Free PMC article. Updated. Preprint.
References
-
- Siebel C, Lendahl U. Notch signaling in development, tissue homeostasis, and disease. Physiol Rev. 2017;97(4):1235–1294. - PubMed
-
- Weng AP, Ferrando AA, Lee W, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306(5694):269–271. - PubMed
-
- Krop I, Demuth T, Guthrie T, et al. Phase I pharmacologic and pharmacodynamic study of the gamma secretase (Notch) inhibitor MK-0752 in adult patients with advanced solid tumors. J Clin Oncol. 2012;30(19):2307–2313. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

