Cleavage of VAMP2/3 Affects Oligodendrocyte Lineage Development in the Developing Mouse Spinal Cord
- PMID: 37620160
- PMCID: PMC10538588
- DOI: 10.1523/JNEUROSCI.2206-21.2023
Cleavage of VAMP2/3 Affects Oligodendrocyte Lineage Development in the Developing Mouse Spinal Cord
Abstract
In the developing and adult CNS, new oligodendrocytes (OLs) are generated from a population of cells known as oligodendrocyte precursor cells (OPCs). As they begin to differentiate, OPCs undergo a series of highly regulated changes to morphology, gene expression, and membrane organization. This stage represents a critical bottleneck in oligodendrogliogenesis, and the regulatory program that guides it is still not fully understood. Here, we show that in vivo toxin-mediated cleavage of the vesicle associated SNARE proteins VAMP2/3 in the OL lineage of both male and female mice impairs the ability of early OLs to mature into functional, myelinating OLs. In the developing mouse spinal cord, many VAMP2/3-cleaved OLs appeared to stall in the premyelinating, early OL stage, resulting in an overall loss of both myelin density and OL number. The Src kinase Fyn, a key regulator of oligodendrogliogenesis and myelination, is highly expressed among premyelinating OLs, but its expression decreases as OLs mature. We found that OLs with cleaved VAMP2/3 in the spinal cord white matter showed significantly higher expression of Fyn compared with neighboring control cells, potentially because of an extended premyelinating stage. Overall, our results show that functional VAMP2/3 in OL lineage cells is essential for proper myelin formation and plays a major role in controlling the maturation and terminal differentiation of premyelinating OLs.SIGNIFICANCE STATEMENT The production of mature oligodendrocytes (OLs) is essential for CNS myelination during development, myelin remodeling in adulthood, and remyelination following injury or in demyelinating disease. Before myelin sheath formation, newly formed OLs undergo a series of highly regulated changes during a stage of their development known as the premyelinating, or early OL stage. This stage acts as a critical checkpoint in OL development, and much is still unknown about the dynamic regulatory processes involved. In this study, we show that VAMP2/3, SNARE proteins involved in vesicular trafficking and secretion play an essential role in regulating premyelinating OL development and are required for healthy myelination in the developing mouse spinal cord.
Keywords: Fyn; SNARE; VAMP; myelin; oligodendrocyte precursor; spinal cord.
Copyright © 2023 the authors.
Figures
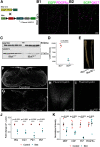

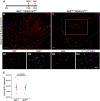
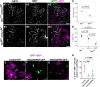
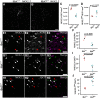



Comment in
-
Vesicle Fusion in Oligodendrocyte Maturation and Myelination.J Neurosci. 2024 Apr 17;44(16):e2203232024. doi: 10.1523/JNEUROSCI.2203-23.2024. J Neurosci. 2024. PMID: 38631918 Free PMC article. No abstract available.
References
-
- Bijlard M, Klunder B, de Jonge JC, Nomden A, Tyagi S, de Vries H, Hoekstra D, Baron W (2015) Transcriptional expression of myelin basic protein in oligodendrocytes depends on functional syntaxin 4: a potential correlation with autocrine signaling. Mol Cell Biol 35:675–687. 10.1128/MCB.01389-14 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
