Circadian clock regulator Bmal1 gates axon regeneration via Tet3 epigenetics in mouse sensory neurons
- PMID: 37620297
- PMCID: PMC10449865
- DOI: 10.1038/s41467-023-40816-7
Circadian clock regulator Bmal1 gates axon regeneration via Tet3 epigenetics in mouse sensory neurons
Abstract
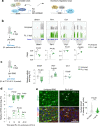
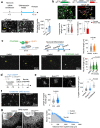
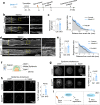
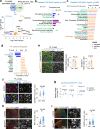
Axon regeneration of dorsal root ganglia (DRG) neurons after peripheral axotomy involves reconfiguration of gene regulatory circuits to establish regenerative gene programs. However, the underlying mechanisms remain unclear. Here, through an unbiased survey, we show that the binding motif of Bmal1, a central transcription factor of the circadian clock, is enriched in differentially hydroxymethylated regions (DhMRs) of mouse DRG after peripheral lesion. By applying conditional deletion of Bmal1 in neurons, in vitro and in vivo neurite outgrowth assays, as well as transcriptomic profiling, we demonstrate that Bmal1 inhibits axon regeneration, in part through a functional link with the epigenetic factor Tet3. Mechanistically, we reveal that Bmal1 acts as a gatekeeper of neuroepigenetic responses to axonal injury by limiting Tet3 expression and restricting 5hmC modifications. Bmal1-regulated genes not only concern axon growth, but also stress responses and energy homeostasis. Furthermore, we uncover an epigenetic rhythm of diurnal oscillation of Tet3 and 5hmC levels in DRG neurons, corresponding to time-of-day effect on axon growth potential. Collectively, our studies demonstrate that targeting Bmal1 enhances axon regeneration.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

