RNAPII-dependent ATM signaling at collisions with replication forks
- PMID: 37620345
- PMCID: PMC10449895
- DOI: 10.1038/s41467-023-40924-4
RNAPII-dependent ATM signaling at collisions with replication forks
Abstract
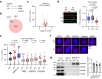
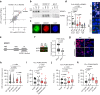
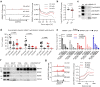
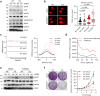
Deregulation of RNA Polymerase II (RNAPII) by oncogenic signaling leads to collisions of RNAPII with DNA synthesis machinery (transcription-replication conflicts, TRCs). TRCs can result in DNA damage and are thought to underlie genomic instability in tumor cells. Here we provide evidence that elongating RNAPII nucleates activation of the ATM kinase at TRCs to stimulate DNA repair. We show the ATPase WRNIP1 associates with RNAPII and limits ATM activation during unperturbed cell cycle. WRNIP1 binding to elongating RNAPII requires catalytic activity of the ubiquitin ligase HUWE1. Mutation of HUWE1 induces TRCs, promotes WRNIP1 dissociation from RNAPII and binding to the replisome, stimulating ATM recruitment and activation at RNAPII. TRCs and translocation of WRNIP1 are rapidly induced in response to hydroxyurea treatment to activate ATM and facilitate subsequent DNA repair. We propose that TRCs can provide a controlled mechanism for stalling of replication forks and ATM activation, instrumental in cellular response to replicative stress.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Guha S, Bhaumik SR. Transcription-coupled DNA double-strand break repair. DNA Repair (Amst.) 2022;109:103211. - PubMed
-
- Bermejo R, Lai MS, Foiani M. Preventing replication stress to maintain genome stability: resolving conflicts between replication and transcription. Mol. Cell. 2012;45:710–718. - PubMed
-
- Garcia-Muse T, Aguilera A. R loops: from physiological to pathological roles. Cell. 2019;179:604–618. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

