Structural snapshots along K48-linked ubiquitin chain formation by the HECT E3 UBR5
- PMID: 37620400
- PMCID: PMC10830417
- DOI: 10.1038/s41589-023-01414-2
Structural snapshots along K48-linked ubiquitin chain formation by the HECT E3 UBR5
Abstract
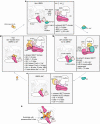
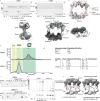
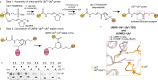
Ubiquitin (Ub) chain formation by homologous to E6AP C-terminus (HECT)-family E3 ligases regulates vast biology, yet the structural mechanisms remain unknown. We used chemistry and cryo-electron microscopy (cryo-EM) to visualize stable mimics of the intermediates along K48-linked Ub chain formation by the human E3, UBR5. The structural data reveal a ≈ 620 kDa UBR5 dimer as the functional unit, comprising a scaffold with flexibly tethered Ub-associated (UBA) domains, and elaborately arranged HECT domains. Chains are forged by a UBA domain capturing an acceptor Ub, with its K48 lured into the active site by numerous interactions between the acceptor Ub, manifold UBR5 elements and the donor Ub. The cryo-EM reconstructions allow defining conserved HECT domain conformations catalyzing Ub transfer from E2 to E3 and from E3. Our data show how a full-length E3, ubiquitins to be adjoined, E2 and intermediary products guide a feed-forward HECT domain conformational cycle establishing a highly efficient, broadly targeting, K48-linked Ub chain forging machine.
© 2023. The Author(s).
Conflict of interest statement
B.A.S. is a member of the scientific advisory boards of Interline Therapeutics and BioTheryX and co-inventor of intellectual property licensed to Cinsano. The remaining authors declare no competing interest.
Figures








References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

