Nivolumab plus ipilimumab in advanced salivary gland cancer: a phase 2 trial
- PMID: 37620627
- PMCID: PMC11293616
- DOI: 10.1038/s41591-023-02518-x
Nivolumab plus ipilimumab in advanced salivary gland cancer: a phase 2 trial
Abstract
Salivary gland cancers (SGCs) are rare, aggressive cancers without effective treatments when metastasized. We conducted a phase 2 trial evaluating nivolumab (nivo, anti-PD-1) and ipilimumab (ipi, anti-CTLA-4) in 64 patients with metastatic SGC enrolled in two histology-based cohorts (32 patients each): adenoid cystic carcinoma (ACC; cohort 1) and other SGCs (cohort 2). The primary efficacy endpoint (≥4 objective responses) was met in cohort 2 (5/32, 16%) but not in cohort 1 (2/32, 6%). Treatment safety/tolerability and progression-free survival (PFS) were secondary endpoints. Treatment-related adverse events grade ≥3 occurred in 24 of 64 (38%) patients across both cohorts, and median PFS was 4.4 months (95% confidence interval (CI): 2.4, 8.3) and 2.2 months (95% CI: 1.8, 5.3) for cohorts 1 and 2, respectively. We present whole-exome, RNA and T cell receptor (TCR) sequencing data from pre-treatment and on-treatment tumors and immune cell flow cytometry and TCR sequencing from peripheral blood at serial timepoints. Responding tumors universally demonstrated clonal expansion of pre-existing T cells and mutational contraction. Responding ACCs harbored neoantigens, including fusion-derived neoepitopes, that induced T cell responses ex vivo. This study shows that nivo+ipi has limited efficacy in ACC, albeit with infrequent, exceptional responses, and that it could be promising for non-ACC SGCs, particularly salivary duct carcinomas. ClinicalTrials.gov identifier: NCT03172624 .
© 2023. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
A.L.H. reports research funding for clinical trials from Allos Therapeutics, Astellas Pharma, AstraZeneca, Bayer, Ayala Pharmaceuticals, Bristol Myers Squibb, Genentech, Celldex Therapeutics, Daiichi Sankyo, Eisai., Elevar Therapeutics, Eli Lilly & Company, Genentech/Roche, Hoikpia, Kolltan Pharmaceuticals, Kura Oncology, Merck, Novartis, Pfizer, Poseida and Verastem; service in consulting/advisory roles for AffyImmune Therapeutics, AstraZeneca, Ayala Pharmaceuticals, Bristol Myers Squibb, Cellestia Biotech, Coherus, CureVac, Eisai, Elevar Therapeutics, Exelixis, Expert Conncet, Genzyme, InxMed, Kura Oncology, McGivney Global Advisors, Merck, Novartis, CureVac, Prelude Therapeutics, Regeneron, Rgenta, Remix Therapeutics, Sanofi, Sun Pharma, the Chemotherapy Foundation and TRM Oncology; service on speakers’ bureaus for Medscape, Omniprex America, Novartis and Physician Education Resource; and receipt of travel/accommodations expenses from Janssen Oncology, Merck, Kura Oncology, Ignyta, Ayala Pharmaceuticals and KLUS Pharma, outside the submitted work. A.L.H. is also inventor on a patent for the use of lesional dosimetry methods for tailoring targeted radiotherapy in cancer. L.G.T.M. is listed as an inventor on intellectual property held by MSK on using tumor mutation burden to predict immunotherapy response, with pending patent, which has been licensed to Personal Genome Diagnostics. T.A.C. reports, all outside the submitted work, being a co-founder of Gritstone Oncology and holding equity; holding equity in An2H and acknowledging grant funding from Bristol Myers Squibb, AstraZeneca, Illumina, Pfizer, An2H and Eisai; having served as an advisor for Bristol Myers, MedImmune, Squibb, Illumina, Eisai, AstraZeneca and An2H; andd being an inventor on intellectual property held by MSK on using tumor mutation burden to predict immunotherapy response, with pending patent, which has been licensed to Personal Genome Diagnostics. C.A.K. is a scientific co-founder and equity holder of Affini-T Therapeutics; is a compensated member of the scientific and/or clinical advisory boards for Achilles Therapeutics, Affini-T Therapeutics, Aleta BioTherapeutics, Bellicum Pharmaceuticals, Catamaran Bio, Obsidian Therapeutics and T-knife; has consulted for Bristol Myers Squibb, Decheng Capital, PACT Pharma and Roche/Genentech; and has patents broadly related to cell and gene therapy outside the scope of this work. N.R. reports research funding from ArcherDx and Repare Therapeutics and personal fees from Illumina, PaigeAI and Pfizer Canada, outside the submitted work. E.J.S. reports institutional research funding from Merck, outside the submitted work, and personal fees from Eli Lilly & Company, Blueprint Medicines Corporation, Regeneron Pharmaceuticals, Loxo Oncology and Eisai, outside the submitted work. L.A.D. reports research funding and personal fees from CUE-101, Eisai, CUE-101, Replimune Group and Regeneron Pharmaceuticals and service on an advisory board at Merck, outside the submitted work. V.T. reports holding stock in Infinity Pharmaceuticals, Bluebird Bio and Mersana Therapeutics. C.L.Z. is linked to investigator-initiated clinical trials in collaboration with Bristol Myers Squibb, outside the submitted work. D.G.P. reports grants from the National Institutes of Health (NIH) and the Philanthropy-Serra Fund; research support from Hookipa Pharma; and personal fees from Nykode and Hookipa Pharma, outside the submitted work. B.B. is an employee of AstraZeneca. Z.N. is an employee of PPD, part of Thermo Fisher Scientific. W.Y. is an employee of Eli Lilly & Company. V.M. is listed as an inventor on a patent assigned to MSK broadly related to determinants of cancer response to immunotherapy. J.L.V., S.J., C.W.R.F., F.K., C.Y.H., M.P., N.K., R.M.S., I.O., A.K., L.S.M., J.V.F., K.K.N., J.E. and S.H. declare no competing interests.
Figures




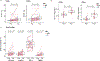

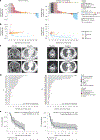



References
-
- Speight PM & Barrett AW Salivary gland tumours. Oral. Dis 8, 229–240 (2002). - PubMed
-
- Bennett AK, Mills SE & Wick MR Salivary-type neoplasms of the breast and lung. Semin. Diagn. Pathol 20, 279–304 (2003). - PubMed
-
- WHO Classification of Head and Neck Tumours 4th edn (eds EI-Naggar AK, Chan JK, Grandis JR, Takata T. & Slootweg P) (IARC, 2017).
-
- Laurie SA, Ho AL, Fury MG, Sherman E. & Pfister DG Systemic therapy in the management of metastatic or locally recurrent adenoid cystic carcinoma of the salivary glands: a systematic review. Lancet Oncol. 12, 815–824 (2010). - PubMed
-
- Laurie SA & Licitra L. Systemic therapy in the palliative management of advanced salivary gland cancers. J. Clin. Oncol 24, 2673–2678 (2006). - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

