Deciphering the genetic landscape of obesity: a data-driven approach to identifying plausible causal genes and therapeutic targets
- PMID: 37620670
- PMCID: PMC10678330
- DOI: 10.1038/s10038-023-01189-3
Deciphering the genetic landscape of obesity: a data-driven approach to identifying plausible causal genes and therapeutic targets
Abstract
Objectives: Genome-wide association studies (GWAS) have successfully revealed numerous susceptibility loci for obesity. However, identifying the causal genes, pathways, and tissues/cell types responsible for these associations remains a challenge, and standardized analysis workflows are lacking. Additionally, due to limited treatment options for obesity, there is a need for the development of new pharmacological therapies. This study aimed to address these issues by performing step-wise utilization of knowledgebase for gene prioritization and assessing the potential relevance of key obesity genes as therapeutic targets.
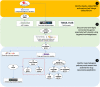
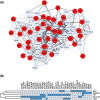
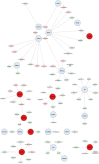
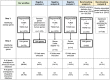
Methods and results: First, we generated a list of 28,787 obesity-associated SNPs from the publicly available GWAS dataset (approximately 800,000 individuals in the GIANT meta-analysis). Then, we prioritized 1372 genes with significant in silico evidence against genomic and transcriptomic data, including transcriptionally regulated genes in the brain from transcriptome-wide association studies. In further narrowing down the gene list, we selected key genes, which we found to be useful for the discovery of potential drug seeds as demonstrated in lipid GWAS separately. We thus identified 74 key genes for obesity, which are highly interconnected and enriched in several biological processes that contribute to obesity, including energy expenditure and homeostasis. Of 74 key genes, 37 had not been reported for the pathophysiology of obesity. Finally, by drug-gene interaction analysis, we detected 23 (of 74) key genes that are potential targets for 78 approved and marketed drugs.
Conclusions: Our results provide valuable insights into new treatment options for obesity through a data-driven approach that integrates multiple up-to-date knowledgebases.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Epigenomic and Transcriptomic Prioritization of Candidate Obesity-Risk Regulatory GWAS SNPs.Int J Mol Sci. 2022 Jan 23;23(3):1271. doi: 10.3390/ijms23031271. Int J Mol Sci. 2022. PMID: 35163195 Free PMC article.
-
Integrative analysis of transcriptome and proteome wide association studies prioritized functional genes for obesity.Hum Genet. 2025 Jan;144(1):31-41. doi: 10.1007/s00439-024-02714-w. Epub 2024 Nov 4. Hum Genet. 2025. PMID: 39495296
-
An integrative systems genetics approach reveals potential causal genes and pathways related to obesity.Genome Med. 2015 Oct 20;7:105. doi: 10.1186/s13073-015-0229-0. Genome Med. 2015. PMID: 26482556 Free PMC article.
-
Strategies to identify causal common genetic variants and corresponding effector genes for paediatric obesity.Pediatr Obes. 2022 Dec;17(12):e12968. doi: 10.1111/ijpo.12968. Epub 2022 Aug 16. Pediatr Obes. 2022. PMID: 35971868 Review.
-
Multivariate analysis of genome-wide data to identify potential pleiotropic genes for type 2 diabetes, obesity and coronary artery disease using MetaCCA.Int J Cardiol. 2019 May 15;283:144-150. doi: 10.1016/j.ijcard.2018.10.102. Epub 2018 Oct 31. Int J Cardiol. 2019. PMID: 30459114 Review.
Cited by
-
The Human Energy Balance: Uncovering the Hidden Variables of Obesity.Diseases. 2025 Feb 13;13(2):55. doi: 10.3390/diseases13020055. Diseases. 2025. PMID: 39997062 Free PMC article.
-
Considerations on efforts needed to improve our understanding of the genetics of obesity.Int J Obes (Lond). 2025 Feb;49(2):206-210. doi: 10.1038/s41366-024-01528-0. Epub 2024 Jun 7. Int J Obes (Lond). 2025. PMID: 38849463 Free PMC article. Review. No abstract available.
-
14-3-3ζ allows for adipogenesis by modulating chromatin accessibility during the early stages of adipocyte differentiation.Mol Metab. 2025 Jul;97:102159. doi: 10.1016/j.molmet.2025.102159. Epub 2025 Apr 28. Mol Metab. 2025. PMID: 40306359 Free PMC article.
-
Integrating Genetic Insights, Technological Advancements, Screening, and Personalized Pharmacological Interventions in Childhood Obesity.Adv Ther. 2025 Jan;42(1):72-93. doi: 10.1007/s12325-024-03057-8. Epub 2024 Nov 13. Adv Ther. 2025. PMID: 39535684 Free PMC article. Review.
-
Antiobesity Pharmacotherapy for Patients With Genetic Obesity Due to Defects in the Leptin-Melanocortin Pathway.Endocr Rev. 2025 May 9;46(3):418-446. doi: 10.1210/endrev/bnaf004. Endocr Rev. 2025. PMID: 39929239 Free PMC article. Review.
References
-
- Keramat SA, Alam K, Rana RH, Chowdhury R, Farjana F, Hashmi R, et al. Obesity and the risk of developing chronic diseases in middle-aged and older adults: Findings from an Australian longitudinal population survey, 2009–2017. PLoS One. 2021;16:e0260158. doi: 10.1371/journal.pone.0260158. - DOI - PMC - PubMed
-
- Hales CM, National Center for Health S. Prevalence of obesity and severe obesity among adults: United States, 2017–2018. Hyattsville, MD: U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, National Center for Health Statistics; 2020.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
