Chromosome-level genome assembly of Babesia caballi reveals diversity of multigene families among Babesia species
- PMID: 37620766
- PMCID: PMC10463595
- DOI: 10.1186/s12864-023-09540-w
Chromosome-level genome assembly of Babesia caballi reveals diversity of multigene families among Babesia species
Abstract
Background: Babesia caballi is an intraerythrocytic parasite from the phylum Apicomplexa, capable of infecting equids and causing equine piroplasmosis. However, since there is limited genome information available on B. caballi, molecular mechanisms involved in host specificity and pathogenicity of this species have not been fully elucidated yet.
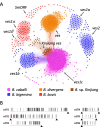
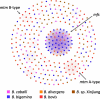
Results: Genomic DNA from a B. caballi subclone was purified and sequenced using both Illumina and Nanopore technologies. The resulting assembled sequence consisted of nine contigs with a size of 12.9 Mbp, rendering a total of 5,910 protein-coding genes. The phylogenetic tree of Apicomplexan species was reconstructed using 263 orthologous genes. We identified 481 ves1-like genes and named "ves1c". In contrast, expansion of the major facilitator superfamily (mfs) observed in closely related B. bigemina and B. ovata species was not found in B. caballi. A set of repetitive units containing an open reading frame with a size of 297 bp was also identified.
Conclusions: We present a chromosome-level genome assembly of B. caballi. Our genomic data may contribute to estimating gene expansion events involving multigene families and exploring the evolution of species from this genus.
Keywords: Babesia caballi; Comparative genomics; Equine babesiosis; Multigene expansion.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

