Understanding the underlying genetic mechanisms for age at first calving, inter-calving period and scrotal circumference in Bonsmara cattle
- PMID: 37620802
- PMCID: PMC10464233
- DOI: 10.1186/s12864-023-09518-8
Understanding the underlying genetic mechanisms for age at first calving, inter-calving period and scrotal circumference in Bonsmara cattle
Abstract
Background: Reproduction is a key feature of the sustainability of a species and thus represents an important component in livestock genetic improvement programs. Most reproductive traits are lowly heritable. In order to gain a better understanding of the underlying genetic basis of these traits, a genome-wide association was conducted for age at first calving (AFC), first inter-calving period (ICP) and scrotal circumference (SC) within the South African Bonsmara breed. Phenotypes and genotypes (120,692 single nucleotide polymorphisms (SNPs) post editing) were available on 7,128 South African Bonsmara cattle; the association analyses were undertaken using linear mixed models.
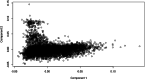
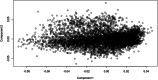
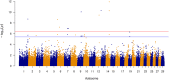
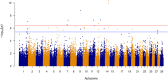
Results: Genomic restricted maximum likelihood analysis of the 7,128 SA Bonsmara cattle yielded genomic heritability's of 0.183 (SE = 0.021) for AFC, 0.207 (SE = 0.022) for ICP and 0.209 (SE = 0.019) for SC. A total of 16, 23 and 51 suggestive (P ≤ 4 × 10-6) SNPs were associated with AFC, ICP and SC, while 11, 11 and 44 significant (P ≤ 4 × 10-7) SNPs were associated with AFC, ICP and SC respectively. A total of 11 quantitative trait loci (QTL) and 11 candidate genes were co-located with these associated SNPs for AFC, with 10 QTL harbouring 11 candidate genes for ICP and 41 QTL containing 40 candidate genes for SC. The QTL identified were close to genes previously associated with carcass, fertility, growth and milk-related traits. The biological pathways influenced by these genes include carbohydrate catabolic processes, cellular development, iron homeostasis, lipid metabolism and storage, immune response, ovarian follicle development and the regulation of DNA transcription and RNA translation.
Conclusions: This was the first attempt to study the underlying polymorphisms associated with reproduction in South African beef cattle. Genes previously reported in cattle breeds for numerous traits bar AFC, ICP or SC were detected in this study. Over 20 different genes have not been previously reported in beef cattle populations and may have been associated due to the unique genetic composite background of the SA Bonsmara breed.
Keywords: Bioinformatics; Deregression; Estimated breeding values; Genomics; Regression; Single nucleotide polymorphism.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
I declare that the authors have no competing interests as defined by BMC, or other interests that might be perceived to influence the results and/or discussion reported in this paper.
Figures

References
-
- Grobler SM, Scholtz MM, Greyling JPC, Neser FWC. Reproduction performance of beef cattle mated naturally following synchronization in the Central Bushveld bioregion of South Africa. S Afr J Anim Sci. 2014;44:70–4.
-
- Bonsma J. Livestock production - a global approach. Cape Town, South Africa: Tafelberg Publishers Ltd; 1983.
-
- Meyer K, Hammond K, Parnell PF, MacKinnon MJ, Sivarajasingam S. Estimates of heritability and repeatability for reproductive traits in australian beef cattle. Livest Prod Sci. 1990;25:15–30.
MeSH terms
LinkOut - more resources
Full Text Sources

