Seedability: optimizing alignment parameters for sensitive sequence comparison
- PMID: 37621456
- PMCID: PMC10444664
- DOI: 10.1093/bioadv/vbad108
Seedability: optimizing alignment parameters for sensitive sequence comparison
Abstract
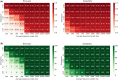
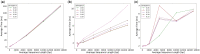
Motivation: Most sequence alignment techniques make use of exact k-mer hits, called seeds, as anchors to optimize alignment speed. A large number of bioinformatics tools employing seed-based alignment techniques, such as , use a single value of k per sequencing technology, without a strong guarantee that this is the best possible value. Given the ubiquity of sequence alignment, identifying values of k that lead to more sensitive alignments is thus an important task. To aid this, we present , a seed-based alignment framework designed for estimating an optimal seed k-mer length (as well as a minimal number of shared seeds) based on a given alignment identity threshold. In particular, we were motivated to make more sensitive in the pairwise alignment of short sequences.
Results: The experimental results herein show improved alignments of short and divergent sequences when using the parameter values determined by in comparison to the default values of . We also show several cases of pairs of real divergent sequences, where the default parameter values of yield no output alignments, but the values output by produce plausible alignments.
Availability and implementation: https://github.com/lorrainea/Seedability (distributed under GPL v3.0).
© The Author(s) 2023. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures


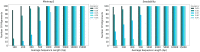

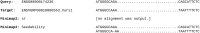
References
-
- Altschul SF, Gish W, Miller W. et al. Basic local alignment search tool. J Mol Biol 1990;215:403–10. - PubMed
-
- Charalampopoulos P, Crochemore M, Fici G. et al. Alignment-free sequence comparison using absent words. Inf Comput 2018;262:57–68.
-
- Chikhi R, Medvedev P.. Informed and automated k-mer size selection for genome assembly. Bioinformatics 2013;30:31–7. - PubMed
-
- Dewey CN. 2012. Whole-genome alignment. In: Anisimova M. (ed.), Evolutionary Genomics: Statistical and Computational Methods, Vol. 1. Totowa, NJ: Humana Press, 237–57.
LinkOut - more resources
Full Text Sources
Miscellaneous

