Emergence of a new genotype of clade 2.3.4.4b H5N1 highly pathogenic avian influenza A viruses in Bangladesh
- PMID: 37622753
- PMCID: PMC10563617
- DOI: 10.1080/22221751.2023.2252510
Emergence of a new genotype of clade 2.3.4.4b H5N1 highly pathogenic avian influenza A viruses in Bangladesh
Abstract
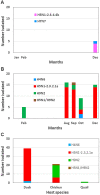
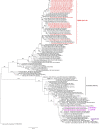
Influenza virological surveillance was conducted in Bangladesh from January to December 2021 in live poultry markets (LPMs) and in Tanguar Haor, a wetland region where domestic ducks have frequent contact with migratory birds. The predominant viruses circulating in LPMs were low pathogenic avian influenza (LPAI) H9N2 and clade 2.3.2.1a highly pathogenic avian influenza (HPAI) H5N1 viruses. Additional LPAIs were found in both LPM (H4N6) and Tanguar Haor wetlands (H7N7). Genetic analyses of these LPAIs strongly suggested long-distance movement of viruses along the Central Asian migratory bird flyway. We also detected a novel clade 2.3.4.4b H5N1 virus from ducks in free-range farms in Tanguar Haor that was similar to viruses first detected in October 2020 in The Netherlands but with a different PB2. Identification of clade 2.3.4.4b HPAI H5N1 viruses in Tanguar Haor provides continued support of the role of migratory birds in transboundary movement of influenza A viruses (IAV), including HPAI viruses. Domestic ducks in free range farm in wetland areas, like Tangua Haor, serve as a conduit for the introduction of LPAI and HPAI viruses into Bangladesh. Clade 2.3.4.4b viruses have dominated in many regions of the world since mid-2021, and it remains to be seen if these viruses will replace the endemic clade 2.3.2.1a H5N1 viruses in Bangladesh.
Keywords: Bangladesh; Surveillance; Tanguar Haor; avian influenza A virus; clade 2.3.4.4b H5N1; domestic ducks; migratory birds; reassortment.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures



References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
