Study of SARS-CoV-2 Spike Protein Wild-Type and the Variants of Concern Real-Time Interactions with Monoclonal Antibodies and Convalescent Human Serum
- PMID: 37622870
- PMCID: PMC10452135
- DOI: 10.3390/bios13080784
Study of SARS-CoV-2 Spike Protein Wild-Type and the Variants of Concern Real-Time Interactions with Monoclonal Antibodies and Convalescent Human Serum
Abstract
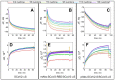
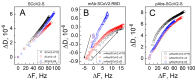
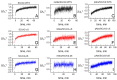
The spike (S) protein and its receptor-binding domain (RBD) of the coronavirus SARS-CoV-2 have been continually evolving, yielding the majority of significant missense mutations and new variants of concern. In this study, we examined how monoclonal antibodies against RBD (mAbs-SCoV2-RBD) and polyclonal antibodies present in convalescent human serum specifically interact with the S protein of wild-type and SARS-CoV-2 variants of concern (VOCs) in real time and how this can be reflected through surface mass density. Moreover, we combined two distinct, label-free measurement techniques: one based on changes in surface electromagnetic waves after reflection from the surface, and the other on changes in acoustic waves. The results demonstrated that dry surface mass density (ΓSE) of mAbs-SCoV2-RBD attached to the RBD of the S protein decreases three-fold, from 148 ng/cm2 to 46 ng/cm2, due to the B.1.351 or so-called beta mutation of coronavirus and its S protein (SCoV2-β). Consequently, the obtained wet mass ΓQCM-D resulted in values two times lower, from 319 ng/cm2 to 158 ng/cm2, and the hydration of mAbs-SCoV2-RBD/SCoV2-β immune complex was 70.88%. Conversely, when polyclonal antibodies present in convalescent human serum form immune complexes with the S protein of SARS-CoV-2 variants of concern, the ΓSE decreased from 279 ng/cm2 to 249 ng/cm2, and ΓQCM-D from 1545 ng/cm2 to 1366 ng/cm2. These results can give insights into the differences between the interaction of monoclonal and polyclonal antibodies with SARS-CoV-2 VOCs.
Keywords: SARS-CoV-2; immunosensor; kinetics; quartz crystal microbalance with dissipation; spectroscopic ellipsometry.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Plikusiene I., Maciulis V., Juciute S., Ramanavicius A., Balevicius Z., Slibinskas R., Kucinskaite-Kodze I., Simanavicius M., Balevicius S., Ramanaviciene A. Investigation of SARS-CoV-2 nucleocapsid protein interaction with a specific antibody by combined spectroscopic ellipsometry and quartz crystal microbalance with dissipation. J. Colloid Interface Sci. 2022;626:113–122. doi: 10.1016/j.jcis.2022.06.119. - DOI - PMC - PubMed
-
- Jerabek-Willemsen M., André T., Wanner R., Roth H.M., Duhr S., Baaske P., Breitsprecher D. MicroScale Thermophoresis: Interaction analysis and beyond. J. Mol. Struct. 2014;1077:101–113. doi: 10.1016/j.molstruc.2014.03.009. - DOI
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

