Length-Dependent Translation Efficiency of ER-Destined Proteins
- PMID: 37623244
- PMCID: PMC10453119
- DOI: 10.3390/cimb45080425
Length-Dependent Translation Efficiency of ER-Destined Proteins
Abstract
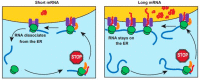
Gene expression is a fundamental process that enables cells to produce specific proteins in a timely and spatially dependent manner. In eukaryotic cells, the complex organization of the cell body requires precise control of protein synthesis and localization. Certain mRNAs encode proteins with an N-terminal signal sequences that direct the translation apparatus toward a specific organelle. Here, we focus on the mechanisms governing the translation of mRNAs, which encode proteins with an endoplasmic reticulum (ER) signal in human cells. The binding of a signal-recognition particle (SRP) to the translation machinery halts protein synthesis until the mRNA-ribosome complex reaches the ER membrane. The commonly accepted model suggests that mRNA that encodes a protein that contains an ER signal peptide continuously repeats the cycle of SRP binding followed by association and dissociation with the ER. In contrast to the current view, we show that the long mRNAs remain on the ER while being translated. On the other hand, due to low ribosome occupancy, the short mRNAs continue the cycle, always facing a translation pause. Ultimately, this leads to a significant drop in the translation efficiency of small, ER-targeted proteins. The proposed mechanism advances our understanding of selective protein synthesis in eukaryotic cells and provides new avenues to enhance protein production in biotechnological settings.
Keywords: endoplasmic reticulum; mRNA; proteosynthesis; ribosome stalling; signal peptide.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources

