SARS-CoV-2 ORF8 accessory protein is a virulence factor
- PMID: 37623322
- PMCID: PMC10653805
- DOI: 10.1128/mbio.00451-23
SARS-CoV-2 ORF8 accessory protein is a virulence factor
Abstract
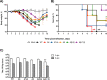
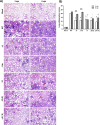
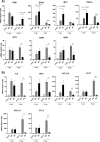
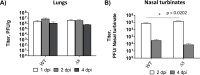
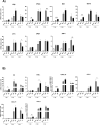
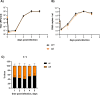
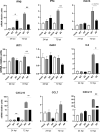
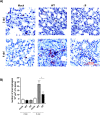
The relevance of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) ORF8 in the pathogenesis of COVID-19 is unclear. Virus natural isolates with deletions in ORF8 were associated with wild milder disease, suggesting that ORF8 might contribute to SARS-CoV-2 virulence. This manuscript shows that ORF8 is involved in inflammation and in the activation of macrophages in two experimental systems: humanized K18-hACE2 transgenic mice and organoid-derived human airway cells. These results identify ORF8 protein as a potential target for COVID-19 therapies.
Keywords: SARS-CoV-2; accessory proteins; coronavirus; viral pathogenesis; virulence.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 588:270–273. doi:10.1038/s41586-020-2951-z - DOI - PMC - PubMed
-
- Gorbalenya AE, Baker SC, Baric RS, de Groot RJ, Drosten C, Gulyaeva AA, Haagmans BL, Lauber C, Leontovich AM, Neuman BW, Penzar D, Perlman S, Poon LLM, Samborskiy DV, Sidorov IA, Sola I, Ziebuhr J, Coronaviridae Study Group of the International Committee on Taxonomy of Viruses . 2020. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol 5:536–544. doi:10.1038/s41564-020-0695-z - DOI - PMC - PubMed
-
- Menachery VD, Mitchell HD, Cockrell AS, Gralinski LE, Yount BL, Graham RL, McAnarney ET, Douglas MG, Scobey T, Beall A, Dinnon K, Kocher JF, Hale AE, Stratton KG, Waters KM, Baric RS, Racaniello VR. 2017. MERS-CoV accessory ORFs play key role for infection and pathogenesis. mBio 8:e00665-17. doi:10.1128/mBio.00665-17 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous
