Copy Number Variations and Gene Mutations Identified by Multiplex Ligation-Dependent Probe Amplification in Romanian Chronic Lymphocytic Leukemia Patients
- PMID: 37623489
- PMCID: PMC10455273
- DOI: 10.3390/jpm13081239
Copy Number Variations and Gene Mutations Identified by Multiplex Ligation-Dependent Probe Amplification in Romanian Chronic Lymphocytic Leukemia Patients
Abstract
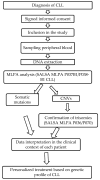
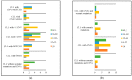
Chronic lymphocytic leukemia (CLL) is known for its wide-ranging clinical and genetic diversity. The study aimed to assess the associations between copy number variations (CNVs) and various biological and clinical features, as well as the survival rates of CLL patients and to evaluate the effectiveness of the multiplex ligation-dependent probe amplification (MLPA) technique in CLL patients.DNA was extracted from 110 patients, and MLPA was performed. Mutations in NOTCH1, SF3B1, and MYD88 were also analyzed. A total of 52 patients showed at least one CNV, 26 had at least one somatic mutation, and 10 presented both, CNVs, and somatic mutations. The most commonly identified CNVs were del(114.3), del(11q22.3), and dup(12q23.2). Other CNVs identified included del(17p13.1), del(14q32.33), dup(10q23.31), and del(19p13.2). One patient was identified with concomitant trisomy 12, 13, and 19. NOTCH1 and SF3B1 mutations were found in 13 patients each, either alone or in combination with other mutations or CNVs, while MYD88 mutation was identified in one patient. Forty-two patients had normal results. Associations between the investigated CNVs and gene mutations and patients' overall survival were found. The presence of NOTCH1 and SF3B1 mutations or the combination of NOTCH1 mutation and CNVs significantly influenced the survival of patients with CLL. Both mutations are frequently associated with different CNVs. Del(13q) is associated with the longest survival rate, while the shortest survival is found in patients with del(17p). Even if MLPA has constraints, it may be used as the primary routine analysis in patients with CLL.
Keywords: CLL; CNV; MLPA; MYD88; NOTCH1; SF3B1.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Genomic alterations in chronic lymphocytic leukemia and their correlation with clinico-hematological parameters and disease progression.Blood Res. 2020 Sep 30;55(3):131-138. doi: 10.5045/br.2020.2020080. Blood Res. 2020. PMID: 32747613 Free PMC article.
-
Comprehensive chronic lymphocytic leukemia diagnostics by combined multiplex ligation dependent probe amplification (MLPA) and interphase fluorescence in situ hybridization (iFISH).Mol Cytogenet. 2014 Nov 19;7(1):79. doi: 10.1186/s13039-014-0079-2. eCollection 2014. Mol Cytogenet. 2014. PMID: 25435911 Free PMC article.
-
NOTCH1, SF3B1, MDM2 and MYD88 mutations in patients with chronic lymphocytic leukemia.Oncol Lett. 2019 Apr;17(4):4016-4023. doi: 10.3892/ol.2019.10048. Epub 2019 Feb 19. Oncol Lett. 2019. PMID: 30930998 Free PMC article.
-
Perspectives on Precision Medicine in Chronic Lymphocytic Leukemia: Targeting Recurrent Mutations-NOTCH1, SF3B1, MYD88, BIRC3.J Clin Med. 2021 Aug 22;10(16):3735. doi: 10.3390/jcm10163735. J Clin Med. 2021. PMID: 34442029 Free PMC article. Review.
-
Therapeutic Options for Patients with TP53 Deficient Chronic Lymphocytic Leukemia: Narrative Review.Cancer Manag Res. 2021 Feb 12;13:1459-1476. doi: 10.2147/CMAR.S283903. eCollection 2021. Cancer Manag Res. 2021. PMID: 33603488 Free PMC article. Review.
Cited by
-
Evaluation of the Copy Number Variants and Single-Nucleotide Polymorphisms of ABCA3 in Newborns with Respiratory Distress Syndrome-A Pilot Study.Medicina (Kaunas). 2024 Feb 29;60(3):419. doi: 10.3390/medicina60030419. Medicina (Kaunas). 2024. PMID: 38541145 Free PMC article.
-
Analysis of Mutational Status of IGHV, and Cytokine Polymorphisms as Prognostic Factors in Chronic Lymphocytic Leukemia: The Romanian Experience.Int J Mol Sci. 2024 Feb 1;25(3):1799. doi: 10.3390/ijms25031799. Int J Mol Sci. 2024. PMID: 38339076 Free PMC article.
References
-
- Campo E., Cymbalista F., Ghia P., Jäger U., Pospisilova S., Rosenquist R., Schuh A., Stilgenbauer S. TP53 Aberrations in Chronic Lymphocytic Leukemia: An Overview of the Clinical Implications of Improved Diagnostics. Haematologica. 2018;103:1956–1968. doi: 10.3324/haematol.2018.187583. - DOI - PMC - PubMed
-
- Puiggros A., Ramos-Campoy S., Kamaso J., de la Rosa M., Salido M., Melero C., Rodríguez-Rivera M., Bougeon S., Collado R., Gimeno E., et al. Optical Genome Mapping: A Promising New Tool to Assess Genomic Complexity in Chronic Lymphocytic Leukemia (CLL) Cancers. 2022;14:3376. doi: 10.3390/cancers14143376. - DOI - PMC - PubMed
-
- Stauder R., Eichhorst B., Hamaker M.E., Kaplanov K., Morrison V.A., Österborg A., Poddubnaya I., Woyach J.A., Shanafelt T., Smolej L., et al. Management of Chronic Lymphocytic Leukemia (CLL) in the Elderly: A Position Paper from an International Society of Geriatric Oncology (SIOG) Task Force. Ann. Oncol. 2017;28:218–227. doi: 10.1093/annonc/mdw547. - DOI - PubMed
LinkOut - more resources
Full Text Sources

