Proteomic and Transcriptomic Analyses to Decipher the Chitinolytic Response of Jeongeupia spp
- PMID: 37623729
- PMCID: PMC10455584
- DOI: 10.3390/md21080448
Proteomic and Transcriptomic Analyses to Decipher the Chitinolytic Response of Jeongeupia spp
Abstract
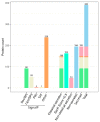
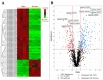
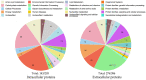
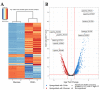
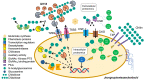
In nature, chitin, the most abundant marine biopolymer, does not accumulate due to the action of chitinolytic organisms, whose saccharification systems provide instructional blueprints for effective chitin conversion. Therefore, discovery and deconstruction of chitinolytic machineries and associated enzyme systems are essential for the advancement of biotechnological chitin valorization. Through combined investigation of the chitin-induced secretome with differential proteomic and transcriptomic analyses, a holistic system biology approach has been applied to unravel the chitin response mechanisms in the Gram-negative Jeongeupia wiesaeckerbachi. Hereby, the majority of the genome-encoded chitinolytic machinery, consisting of various glycoside hydrolases and a lytic polysaccharide monooxygenase, could be detected extracellularly. Intracellular proteomics revealed a distinct translation pattern with significant upregulation of glucosamine transport, metabolism, and chemotaxis-associated proteins. While the differential transcriptomic results suggested the overall recruitment of more genes during chitin metabolism compared to that of glucose, the detected protein-mRNA correlation was low. As one of the first studies of its kind, the involvement of over 350 unique enzymes and 570 unique genes in the catabolic chitin response of a Gram-negative bacterium could be identified through a three-way systems biology approach. Based on the cumulative data, a holistic model for the chitinolytic machinery of Jeongeupia spp. is proposed.
Keywords: chitin; chitinase; chitinolytic; glycosidic hydrolase family 18; lytic polysaccharide monooxygenase; omics; proteomics; transcriptomics.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures
Similar articles
-
The Fish Pathogen Aliivibrio salmonicida LFI1238 Can Degrade and Metabolize Chitin despite Gene Disruption in the Chitinolytic Pathway.Appl Environ Microbiol. 2021 Sep 10;87(19):e0052921. doi: 10.1128/AEM.00529-21. Epub 2021 Sep 10. Appl Environ Microbiol. 2021. PMID: 34319813 Free PMC article.
-
Genomic and Proteomic Study of Andreprevotia ripae Isolated from an Anthill Reveals an Extensive Repertoire of Chitinolytic Enzymes.J Proteome Res. 2021 Aug 6;20(8):4041-4052. doi: 10.1021/acs.jproteome.1c00358. Epub 2021 Jun 30. J Proteome Res. 2021. PMID: 34191517 Free PMC article.
-
Proteomic investigation of the secretome of Cellvibrio japonicus during growth on chitin.Proteomics. 2016 Jul;16(13):1904-14. doi: 10.1002/pmic.201500419. Epub 2016 May 30. Proteomics. 2016. PMID: 27169553
-
Chitinolytic functions in actinobacteria: ecology, enzymes, and evolution.Appl Microbiol Biotechnol. 2018 Sep;102(17):7219-7230. doi: 10.1007/s00253-018-9149-4. Epub 2018 Jun 21. Appl Microbiol Biotechnol. 2018. PMID: 29931600 Free PMC article. Review.
-
Bacterial Chitinase System as a Model of Chitin Biodegradation.Adv Exp Med Biol. 2019;1142:131-151. doi: 10.1007/978-981-13-7318-3_7. Adv Exp Med Biol. 2019. PMID: 31102245 Review.
Cited by
-
High Degree of Polymerization of Chitin Oligosaccharides Produced from Shrimp Shell Waste by Enrichment Microbiota Using Two-Stage Temperature-Controlled Technique of Inducing Enzyme Production and Metagenomic Analysis of Microbiota Succession.Mar Drugs. 2024 Jul 28;22(8):346. doi: 10.3390/md22080346. Mar Drugs. 2024. PMID: 39195462 Free PMC article.
References
-
- Gooday G.W. In: The Ecology of Chitin Degradation BT-Advances in Microbial Ecology. Marshall K.C., editor. Springer; Boston, MA, USA: 1990. pp. 387–430.
-
- Rinaudo M. Chitin and Chitosan: Properties and Applications. Prog. Polym. Sci. 2006;31:603–632. doi: 10.1016/j.progpolymsci.2006.06.001. - DOI
-
- FAO . The State of World Fisheries and Aquaculture 2022. Food & Agriculture Organization; Rome, Italy: 2022.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

