An ARMS-Multiplex PCR Targeting SARS-CoV-2 Omicron Sub-Variants
- PMID: 37623977
- PMCID: PMC10459702
- DOI: 10.3390/pathogens12081017
An ARMS-Multiplex PCR Targeting SARS-CoV-2 Omicron Sub-Variants
Abstract
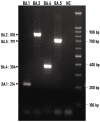
As of November 2021, the SARS-CoV-2 Omicron variant had made its appearance, gradually replacing the predominant Delta variant. Since its emergence, the Omicron variant has been continuously evolving through more than 500 strains, most of which belong to five sub-variants known as BA.1, BA.2, BA.3, BA.4, and BA.5. The aim of this study was to develop a multiplex polymerase chain reaction (PCR) that will be able to distinguish the basic sub-variants of Omicron in a rapid and specific way. Full genome sequences of Omicron strains with high frequency and wide geographical distribution were retrieved by the NCBI Virus and ENA databases. These sequences were compared to each other in order to locate single nucleotide polymorphisms common to all strains of the same sub-variant. These polymorphisms should also be capable of distinguishing Omicron sub-variants not only from each other but from previously circulating variants of SARS-CoV-2 as well. Thus, specific primers targeting characteristic polymorphisms of the four Omicron main branches BA.1, BA.2, BA.4, and BA.5 were designed according to the principles of the amplification refractory mutation system (ARMS) and with the ability to react under multiplex PCR conditions. According to our results, the ARMS-multiplex PCR could successfully distinguish all Omicron sub-variants that carry the corresponding mutations.
Keywords: ARMS; BA.1; BA.2; BA.4; BA.5; Multiplex PCR; Omicron; SARS-CoV-2; variants.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Weekly Epidemiological Update on COVID-19-13 July 2023. Edition 151. [(accessed on 24 July 2023)]. Available online: https://www.who.int/publications/m/item/weekly-epidemiological-update-on....
-
- Sah R., Rais M.A., Mohanty A., Chopra H., Chandran D., Bin Emran T., Dhama K. Omicron (B.1.1.529) variant and its subvariants and lineages may lead to another COVID-19 wave in the world? -An overview of current evidence and counteracting strategies. Int. J. Surg. Open. 2023;55:100625. doi: 10.1016/j.ijso.2023.100625. - DOI - PMC - PubMed
-
- One Year since the Emergence of COVID-19 Virus Variant Omicron. [(accessed on 3 June 2023)]. Available online: https://www.who.int/news-room/feature-stories/detail/one-year-since-the-....
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

