Analysis of Fecal Microbial Changes in Young Calves Following Bovine Rotavirus Infection
- PMID: 37624283
- PMCID: PMC10459456
- DOI: 10.3390/vetsci10080496
Analysis of Fecal Microbial Changes in Young Calves Following Bovine Rotavirus Infection
Abstract
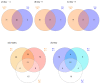
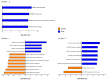
The objective of the present study was to identify changes in fecal microbiota and predict the functional features of healthy calves and those infected with rotavirus over time. Six Holstein calves (average body weight 43.63 ± 1.19 kg, age-matched within 5-7 d) were randomly selected and distributed into two groups which contained three calves each. Fecal samples were taken 3 days before inoculation and on days 1 and 7 post-inoculation. The 16S rRNA gene amplicon sequencing was performed. Bacterial diversity tended to decrease in the rota group, as indicated by the alpha (evenness, p = 0.074 and Shannon, p = 0.055) and beta (Bray-Curtis dissimilarity, p = 0.099) diversity at 1 day post-inoculation. Differences in the bacterial taxa between healthy and rota-infected calves were detected using a linear discriminant analysis effect size (LDA > 2.0, p < 0.05). Rota calves had a higher abundance of certain bacterial taxa, such as Enterococcus, Streptococcus, and Escherichia-Shigella, and a lower abundance of bacteria that contribute to the production of short-chain fatty acids, such as Alistipes, Faecalibacterium, Pseudoflavonifractor, Subdoligranulum, Alloprevotella, Butyricicoccus, and Ruminococcus, compared to the healthy calves. The observed changes in the fecal microbiota of the rota-infected group compared to the healthy group indicated potential dysbiosis. This was further supported by significant differences in the predicted functional metagenomic profiles of these microbial communities. We suggest that calves infected with bovine rotavirus had bacterial dysbiosis, which was characterized by lower diversity and fewer observed genera than the fecal microbiota of healthy calves.
Keywords: bovine rotavirus; fecal microbiota; holstein calves; metataxonomic.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources

