KRASG12D inhibition reprograms the microenvironment of early and advanced pancreatic cancer to promote FAS-mediated killing by CD8+ T cells
- PMID: 37625401
- PMCID: PMC10785700
- DOI: 10.1016/j.ccell.2023.07.002
KRASG12D inhibition reprograms the microenvironment of early and advanced pancreatic cancer to promote FAS-mediated killing by CD8+ T cells
Abstract
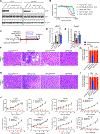
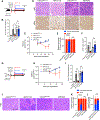
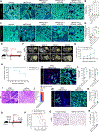
The KRASG12D mutation is present in nearly half of pancreatic adenocarcinomas (PDAC). We investigated the effects of inhibiting the KRASG12D mutant protein with MRTX1133, a non-covalent small molecule inhibitor of KRASG12D, on early and advanced PDAC and its influence on the tumor microenvironment. Employing 16 different models of KRASG12D-driven PDAC, we demonstrate that MRTX1133 reverses early PDAC growth, increases intratumoral CD8+ effector T cells, decreases myeloid infiltration, and reprograms cancer-associated fibroblasts. MRTX1133 leads to regression of both established PanINs and advanced PDAC. Regression of advanced PDAC requires CD8+ T cells and immune checkpoint blockade (ICB) synergizes with MRTX1133 to eradicate PDAC and prolong overall survival. Mechanistically, inhibition of KRASG12D in advanced PDAC and human patient derived organoids induces FAS expression in cancer cells and facilitates CD8+ T cell-mediated death. Collectively, this study provides a rationale for a synergistic combination of MRTX1133 with ICB in clinical trials.
Copyright © 2023 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests A.M. receives royalties for a pancreatic cancer biomarker test from Cosmos Wisdom Biotechnology, and this financial relationship is managed and monitored by the UTMDACC Conflict of Interest Committee. A.M. is also listed as an inventor on a patent that has been licensed by Johns Hopkins University to Thrive Earlier Detection and serves as a consultant for Freenome and Tezcat Biosciences.
Figures




References
-
- Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, Rustgi AK, Chang S, and Tuveson DA (2005). Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell 7, 469–483. 10.1016/j.ccr.2005.04.023. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

