The Genomic Basis of Adaptation to High Elevations in Africanized Honey Bees
- PMID: 37625795
- PMCID: PMC10484329
- DOI: 10.1093/gbe/evad157
The Genomic Basis of Adaptation to High Elevations in Africanized Honey Bees
Abstract
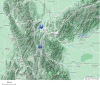
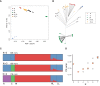
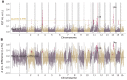
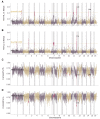
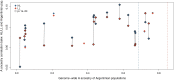
A range of different genetic architectures underpin local adaptation in nature. Honey bees (Apis mellifera) in the Eastern African Mountains harbor high frequencies of two chromosomal inversions that likely govern adaptation to this high-elevation habitat. In the Americas, honey bees are hybrids of European and African ancestries and adaptation to latitudinal variation in climate correlates with the proportion of these ancestries across the genome. It is unknown which, if either, of these forms of genetic variation governs adaptation in honey bees living at high elevations in the Americas. Here, we performed whole-genome sequencing of 29 honey bees from both high- and low-elevation populations in Colombia. Analysis of genetic ancestry indicated that both populations were predominantly of African ancestry, but the East African inversions were not detected. However, individuals in the higher elevation population had significantly higher proportions of European ancestry, likely reflecting local adaptation. Several genomic regions exhibited particularly high differentiation between highland and lowland bees, containing candidate loci for local adaptation. Genes that were highly differentiated between highland and lowland populations were enriched for functions related to reproduction and sperm competition. Furthermore, variation in levels of European ancestry across the genome was correlated between populations of honey bees in the highland population and populations at higher latitudes in South America. The results are consistent with the hypothesis that adaptation to both latitude and elevation in these hybrid honey bees are mediated by variation in ancestry at many loci across the genome.
Keywords: admixture; honey bee; introgression; local adaptation; natural selection.
© The Author(s) 2023. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures
References
-
- Barghi N, Hermisson J, Schlötterer C. 2020. Polygenic adaptation: a unifying framework to understand positive selection. Nat Rev Genet. 21:769–781. - PubMed
-
- Barrier M, Baldwin BG, Robichaux RH, Purugganan MD. 1999. Interspecific hybrid ancestry of a plant adaptive radiation: allopolyploidy of the Hawaiian silversword alliance (Asteraceae) inferred from floral homeotic gene duplications. Mol Biol Evol. 16:1105–1113. - PubMed

