Biochemical characterization of mRNA capping enzyme from Faustovirus
- PMID: 37625853
- PMCID: PMC10578482
- DOI: 10.1261/rna.079738.123
Biochemical characterization of mRNA capping enzyme from Faustovirus
Abstract
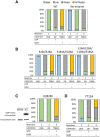
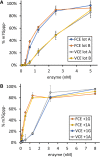
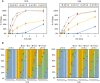
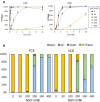
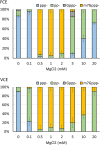
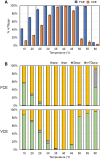
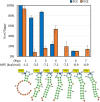
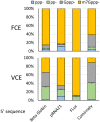
The mammalian mRNA 5' cap structures play important roles in cellular processes such as nuclear export, efficient translation, and evading cellular innate immune surveillance and regulating 5'-mediated mRNA turnover. Hence, installation of the proper 5' cap is crucial in therapeutic applications of synthetic mRNA. The core 5' cap structure, Cap-0, is generated by three sequential enzymatic activities: RNA 5' triphosphatase, RNA guanylyltransferase, and cap N7-guanine methyltransferase. Vaccinia virus RNA capping enzyme (VCE) is a heterodimeric enzyme that has been widely used in synthetic mRNA research and manufacturing. The large subunit of VCE D1R exhibits a modular structure where each of the three structural domains possesses one of the three enzyme activities, whereas the small subunit D12L is required to activate the N7-guanine methyltransferase activity. Here, we report the characterization of a single-subunit RNA capping enzyme from an amoeba giant virus. Faustovirus RNA capping enzyme (FCE) exhibits a modular array of catalytic domains in common with VCE and is highly efficient in generating the Cap-0 structure without an activation subunit. Phylogenetic analysis suggests that FCE and VCE are descended from a common ancestral capping enzyme. We found that compared to VCE, FCE exhibits higher specific activity, higher activity toward RNA containing secondary structures and a free 5' end, and a broader temperature range, properties favorable for synthetic mRNA manufacturing workflows.
Keywords: Faustovirus; RNA capping enzymes; RNA caps; mRNA synthesis; vaccinia capping enzyme.
© 2023 Chan et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures


Similar articles
-
Preparation, and enzymatic activity analysis of an engineered capping enzyme.Enzyme Microb Technol. 2025 Aug;188:110640. doi: 10.1016/j.enzmictec.2025.110640. Epub 2025 Apr 1. Enzyme Microb Technol. 2025. PMID: 40188656
-
The mRNA (guanine-7-)methyltransferase domain of the vaccinia virus mRNA capping enzyme. Expression in Escherichia coli and structural and kinetic comparison to the intact capping enzyme.J Biol Chem. 1994 May 27;269(21):14974-81. J Biol Chem. 1994. PMID: 8195132
-
Catalytic activity of vaccinia mRNA capping enzyme subunits coexpressed in Escherichia coli.J Biol Chem. 1990 Jul 15;265(20):11960-6. J Biol Chem. 1990. PMID: 2164022
-
Enzymology of RNA cap synthesis.Wiley Interdiscip Rev RNA. 2010 Jul-Aug;1(1):152-72. doi: 10.1002/wrna.19. Epub 2010 May 25. Wiley Interdiscip Rev RNA. 2010. PMID: 21956912 Free PMC article. Review.
-
mRNA capping: biological functions and applications.Nucleic Acids Res. 2016 Sep 19;44(16):7511-26. doi: 10.1093/nar/gkw551. Epub 2016 Jun 17. Nucleic Acids Res. 2016. PMID: 27317694 Free PMC article. Review.
Cited by
-
A tripartite cell-free translation system to study mammalian translation.Nat Protoc. 2025 Apr 16. doi: 10.1038/s41596-025-01155-7. Online ahead of print. Nat Protoc. 2025. PMID: 40240502 Review.
-
A comparative exploration of mRNA capping enzymes.Biotechnol Notes. 2024 Nov 20;5:165-172. doi: 10.1016/j.biotno.2024.11.005. eCollection 2024. Biotechnol Notes. 2024. PMID: 39649099 Free PMC article.
-
The design, manufacture and LNP formulation of mRNA for research use.Nat Protoc. 2025 Jun 10. doi: 10.1038/s41596-025-01174-4. Online ahead of print. Nat Protoc. 2025. PMID: 40494942 Review.
-
Therapeutic synthetic and natural materials for immunoengineering.Chem Soc Rev. 2024 Feb 19;53(4):1789-1822. doi: 10.1039/d3cs00805c. Chem Soc Rev. 2024. PMID: 38170619 Free PMC article. Review.
-
IFIT1 is rapidly evolving and exhibits disparate antiviral activities across 11 mammalian orders.bioRxiv [Preprint]. 2025 Jul 1:2024.05.13.593954. doi: 10.1101/2024.05.13.593954. bioRxiv. 2025. PMID: 38798375 Free PMC article. Preprint.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
