Microglial REV-ERBα regulates inflammation and lipid droplet formation to drive tauopathy in male mice
- PMID: 37626048
- PMCID: PMC10457319
- DOI: 10.1038/s41467-023-40927-1
Microglial REV-ERBα regulates inflammation and lipid droplet formation to drive tauopathy in male mice
Erratum in
-
Author Correction: Microglial REV-ERBα regulates inflammation and lipid droplet formation to drive tauopathy in male mice.Nat Commun. 2023 Nov 27;14(1):7760. doi: 10.1038/s41467-023-43360-6. Nat Commun. 2023. PMID: 38012192 Free PMC article. No abstract available.
Abstract
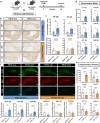
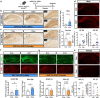
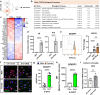
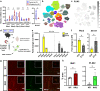
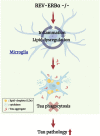
Alzheimer's disease, the most common age-related neurodegenerative disease, is characterized by tau aggregation and associated with disrupted circadian rhythms and dampened clock gene expression. REV-ERBα is a core circadian clock protein which also serves as a nuclear receptor and transcriptional repressor involved in lipid metabolism and macrophage function. Global REV-ERBα deletion has been shown to promote microglial activation and mitigate amyloid plaque formation. However, the cell-autonomous effects of microglial REV-ERBα in healthy brain and in tauopathy are unexplored. Here, we show that microglial REV-ERBα deletion enhances inflammatory signaling, disrupts lipid metabolism, and causes lipid droplet (LD) accumulation specifically in male microglia. These events impair microglial tau phagocytosis, which can be partially rescued by blockage of LD formation. In vivo, microglial REV-ERBα deletion exacerbates tau aggregation and neuroinflammation in two mouse tauopathy models, specifically in male mice. These data demonstrate the importance of microglial lipid droplets in tau accumulation and reveal REV-ERBα as a therapeutically accessible, sex-dependent regulator of microglial inflammatory signaling, lipid metabolism, and tauopathy.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

