Refinement of an ovine-based immunoglobulin therapy against SARS-CoV-2, with comparison of whole IgG versus F(ab')2 fragments
- PMID: 37626085
- PMCID: PMC10457378
- DOI: 10.1038/s41598-023-40277-4
Refinement of an ovine-based immunoglobulin therapy against SARS-CoV-2, with comparison of whole IgG versus F(ab')2 fragments
Erratum in
-
Publisher Correction: Refinement of an ovine-based immunoglobulin therapy against SARS-CoV-2, with comparison of whole IgG versus F(ab')2 fragments.Sci Rep. 2023 Sep 18;13(1):15419. doi: 10.1038/s41598-023-42526-y. Sci Rep. 2023. PMID: 37723274 Free PMC article. No abstract available.
Abstract
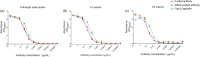
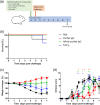
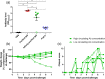
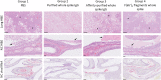
The development of new therapies against SARS-CoV-2 is required to extend the toolkit of intervention strategies to combat the global pandemic. In this study, hyperimmune plasma from sheep immunised with whole spike SARS-CoV-2 recombinant protein has been used to generate candidate products. In addition to purified IgG, we have refined candidate therapies by removing non-specific IgG via affinity binding along with fragmentation to eliminate the Fc region to create F(ab')2 fragments. These preparations were evaluated for in vitro activity and demonstrated to be strongly neutralising against a range of SARS-CoV-2 strains, including Omicron B2.2. In addition, their protection against disease manifestations and viral loads were assessed using a hamster SARS-CoV-2 infection model. Results demonstrated protective effects of both IgG and F(ab')2, with the latter requiring sequential dosing to maintain in vivo activity due to rapid clearance from the circulation.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Sandra Smith and Neville Pope are employees of International Therapeutic Proteins Ltd. Matthew Aldridge is an employee of MicroPharm Ltd. Gareth Humphries and Holger Schuhmann are employees of the Native Antigen Company. All other authors declare no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



References
-
- Findlay-Wilson S, Easterbrook L, Smith S, Pope N, Humphries G, Schuhmann H, et al. Development of a cost-effective ovine antibody-based therapy against SARS-CoV-2 infection and contribution of antibodies specific to the spike subunit proteins. Antiviral Res. 2022;203:105332. doi: 10.1016/j.antiviral.2022.105332. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

