Propensity of selecting mutant parasites for the antimalarial drug cabamiquine
- PMID: 37626093
- PMCID: PMC10457284
- DOI: 10.1038/s41467-023-40974-8
Propensity of selecting mutant parasites for the antimalarial drug cabamiquine
Erratum in
-
Author Correction: Propensity of selecting mutant parasites for the antimalarial drug cabamiquine.Nat Commun. 2023 Sep 6;14(1):5447. doi: 10.1038/s41467-023-41287-6. Nat Commun. 2023. PMID: 37673924 Free PMC article. No abstract available.
Abstract
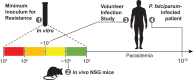
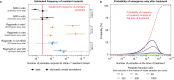
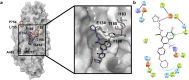
We report an analysis of the propensity of the antimalarial agent cabamiquine, a Plasmodium-specific eukaryotic elongation factor 2 inhibitor, to select for resistant Plasmodium falciparum parasites. Through in vitro studies of laboratory strains and clinical isolates, a humanized mouse model, and volunteer infection studies, we identified resistance-associated mutations at 11 amino acid positions. Of these, six (55%) were present in more than one infection model, indicating translatability across models. Mathematical modelling suggested that resistant mutants were likely pre-existent at the time of drug exposure across studies. Here, we estimated a wide range of frequencies of resistant mutants across the different infection models, much of which can be attributed to stochastic differences resulting from experimental design choices. Structural modelling implicates binding of cabamiquine to a shallow mRNA binding site adjacent to two of the most frequently identified resistance mutations.
© 2023. Springer Nature Limited.
Conflict of interest statement
L.F. is employed by Merck Healthcare, Darmstadt, Germany. C.D.G., C.O., and T.S. are employed by Ares Trading S.A., Eysins, Switzerland, an affiliate of Merck KGaA, Darmstadt, Germany. All other authors declare no competing interest.
Figures


References
-
- World Health Organization. Artemisinin resistance and artemisinin-based combination therapy efficacy: status report. https://www.who.int/docs/default-source/documents/publications/gmp/who-c... (2018).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

