Various arrangements of mobile genetic elements among CC147 subpopulations of Klebsiella pneumoniae harboring blaNDM-1: a comparative genomic analysis of carbapenem resistant strains
- PMID: 37626377
- PMCID: PMC10464136
- DOI: 10.1186/s12929-023-00960-0
Various arrangements of mobile genetic elements among CC147 subpopulations of Klebsiella pneumoniae harboring blaNDM-1: a comparative genomic analysis of carbapenem resistant strains
Abstract
Background: Certain clonal complexes (CCs) of Klebsiella pneumoniae such as CC147 (ST147 and ST392) are major drivers of blaNDM dissemination across the world. ST147 has repeatedly reported from our geographical region, but its population dynamics and evolutionary trajectories need to be further studied.
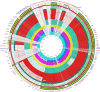
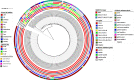
Methods: Comparative genomic analysis of 51 carbapenem-nonsusceptible strains as well as three hypervirulent K. pneumoniae (hvKp) recovered during 16-months of surveillance was performed using various bioinformatics tools. We investigated the genetic proximity of our ST147 strains with publicly available corresponding genomes deposited globally and from neighbor countries in our geographic region.
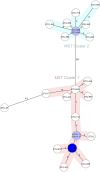
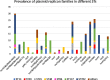
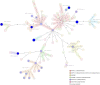
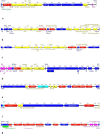
Results: While IncL/M plasmid harboring blaOXA-48 was distributed among divergent clones, blaNDM-1 was circulated by twenty of the 25 CC147 dominant clone and were mostly recovered from the ICU. The NDM-1 core structure was bracketed by a single isoform of mobile genetic elements (MGEs) [ΔISKpn26-NDM-TnAs3-ΔIS3000-Tn5403] and was located on Col440I plasmid in 68.7% of ST392. However, various arrangements of MGEs including MITESen1/MITESen1 composite transposon or combination of MITESen1/ISSen4/IS903B/IS5/ISEhe3 on IncFIb (pB171) were identified in ST147. It seems that ST392 circulated blaNDM-1 in 2018 before being gradually replaced by ST147 from the middle to the end of sample collection in 2019. ST147 strains possessed the highest number of resistance markers and showed high genetic similarity with four public genomes that harbored blaNDM-1 on the same replicon type. Mainly, there was a convergence between clusters and isolated neighboring countries in the minimum-spanning tree. A conserved arrangement of resistance markers/MGEs was linked to methyltransferase armA which was embedded in class 1 integron in 8 isolates of ST147/ST48 high-risk clones.
Conclusion: Our findings highlight the dynamic nature of blaNDM-1 transmission among K. pneumoniae in Iran that occurs both clonally and horizontally via various combinations of MGEs. This is the first analysis of Iranian ST147/NDM + clone in the global context.
Keywords: High-risk clones; Iran; Klebsiella pneumoniae; MGEs; OXA-48; Phylogeny; ST147; Whole genome sequencing; armA.
© 2023. National Science Council of the Republic of China (Taiwan).
Conflict of interest statement
The authors declare none.
Figures

Similar articles
-
The emergence of the hypervirulent Klebsiella pneumoniae (hvKp) strains among circulating clonal complex 147 (CC147) harbouring blaNDM/OXA-48 carbapenemases in a tertiary care center of Iran.Ann Clin Microbiol Antimicrob. 2020 Mar 31;19(1):12. doi: 10.1186/s12941-020-00349-z. Ann Clin Microbiol Antimicrob. 2020. PMID: 32234050 Free PMC article.
-
Molecular characterization of carbapenem-resistant serotype K1 hypervirulent Klebsiella pneumoniae ST11 harbouring blaNDM-1 and blaOXA-48 carbapenemases in Iran.Microb Pathog. 2020 Dec;149:104507. doi: 10.1016/j.micpath.2020.104507. Epub 2020 Sep 17. Microb Pathog. 2020. PMID: 32950637
-
Genomic evolution of the globally disseminated multidrug-resistant Klebsiella pneumoniae clonal group 147.Microb Genom. 2022 Jan;8(1):000737. doi: 10.1099/mgen.0.000737. Microb Genom. 2022. PMID: 35019836 Free PMC article.
-
Molecular Characteristics of an NDM-4 and OXA-181 Co-Producing K51-ST16 Carbapenem-Resistant Klebsiella pneumoniae: Study of Its Potential Dissemination Mediated by Conjugative Plasmids and Insertion Sequences.Antimicrob Agents Chemother. 2023 Jan 24;67(1):e0135422. doi: 10.1128/aac.01354-22. Epub 2023 Jan 5. Antimicrob Agents Chemother. 2023. PMID: 36602346 Free PMC article.
-
Spread of Carbapenem-Resistant Klebsiella pneumoniae Clinical Isolates Producing NDM-Type Metallo-β-Lactamase in Myanmar.Microbiol Spectr. 2022 Aug 31;10(4):e0067322. doi: 10.1128/spectrum.00673-22. Epub 2022 Jun 28. Microbiol Spectr. 2022. PMID: 35762817 Free PMC article.
Cited by
-
Comprehensive genomics reveals novel sequence types of multidrug resistant Klebsiella oxytoca with uncharacterized capsular polysaccharide K- and lipopolysaccharide O-antigen loci from the National Hospital of Uganda.Infect Genet Evol. 2024 Sep;123:105640. doi: 10.1016/j.meegid.2024.105640. Epub 2024 Jul 11. Infect Genet Evol. 2024. PMID: 39002874 Free PMC article.
-
Genomic Characterization of 16S rRNA Methyltransferase-Producing Enterobacterales Reveals the Emergence of Klebsiella pneumoniae ST6260 Harboring rmtF, rmtB, blaNDM-5, blaOXA-232 and blaSFO-1 Genes in a Cancer Hospital in Bulgaria.Antibiotics (Basel). 2024 Oct 10;13(10):950. doi: 10.3390/antibiotics13100950. Antibiotics (Basel). 2024. PMID: 39452216 Free PMC article.
-
First report of coexistence of blaKPC-2 and blaNDM-1 in carbapenem-resistant clinical isolates of Klebsiella aerogenes in Brazil.Front Microbiol. 2024 Feb 14;15:1352851. doi: 10.3389/fmicb.2024.1352851. eCollection 2024. Front Microbiol. 2024. PMID: 38426065 Free PMC article.
-
Genomic Insights into Carbapenem-Resistant Organisms Producing New Delhi Metallo-β-Lactamase in Live Poultry Markets.Microorganisms. 2025 May 23;13(6):1195. doi: 10.3390/microorganisms13061195. Microorganisms. 2025. PMID: 40572083 Free PMC article.
-
High prevalence of carbapenem resistance and clonal expansion of blaNDM gene in Klebsiella pneumoniae isolates in an Iranian referral pediatric hospital.Gut Pathog. 2024 Mar 28;16(1):17. doi: 10.1186/s13099-024-00611-1. Gut Pathog. 2024. PMID: 38549114 Free PMC article.
References
-
- CDC. 2019. Antibiotic resistance threats in the United States, 2019. Atlanta, GA: U.S. Department of Health and Human Services, CDC.
-
- WHO. Antimicrobial Resistance: Global Report on Surveillance 2014. Geneva: World Health Organization; 2014.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

