The endohyphal microbiome: current progress and challenges for scaling down integrative multi-omic microbiome research
- PMID: 37626434
- PMCID: PMC10463477
- DOI: 10.1186/s40168-023-01634-7
The endohyphal microbiome: current progress and challenges for scaling down integrative multi-omic microbiome research
Abstract
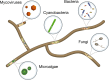
As microbiome research has progressed, it has become clear that most, if not all, eukaryotic organisms are hosts to microbiomes composed of prokaryotes, other eukaryotes, and viruses. Fungi have only recently been considered holobionts with their own microbiomes, as filamentous fungi have been found to harbor bacteria (including cyanobacteria), mycoviruses, other fungi, and whole algal cells within their hyphae. Constituents of this complex endohyphal microbiome have been interrogated using multi-omic approaches. However, a lack of tools, techniques, and standardization for integrative multi-omics for small-scale microbiomes (e.g., intracellular microbiomes) has limited progress towards investigating and understanding the total diversity of the endohyphal microbiome and its functional impacts on fungal hosts. Understanding microbiome impacts on fungal hosts will advance explorations of how "microbiomes within microbiomes" affect broader microbial community dynamics and ecological functions. Progress to date as well as ongoing challenges of performing integrative multi-omics on the endohyphal microbiome is discussed herein. Addressing the challenges associated with the sample extraction, sample preparation, multi-omic data generation, and multi-omic data analysis and integration will help advance current knowledge of the endohyphal microbiome and provide a road map for shrinking microbiome investigations to smaller scales. Video Abstract.
Keywords: Endobacteria; Endohyphal microbiome; Integrative bioinformatics; Multi-omics; Mycovirus.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Challenges, Strategies, and Perspectives for Reference-Independent Longitudinal Multi-Omic Microbiome Studies.Front Genet. 2021 Jun 14;12:666244. doi: 10.3389/fgene.2021.666244. eCollection 2021. Front Genet. 2021. PMID: 34194470 Free PMC article.
-
Unfolding and de-confounding: biologically meaningful causal inference from longitudinal multi-omic networks using METALICA.mSystems. 2024 Oct 22;9(10):e0130323. doi: 10.1128/msystems.01303-23. Epub 2024 Sep 6. mSystems. 2024. PMID: 39240096 Free PMC article.
-
Multi-omic approaches for host-microbiome data integration.Gut Microbes. 2024 Jan-Dec;16(1):2297860. doi: 10.1080/19490976.2023.2297860. Epub 2024 Jan 2. Gut Microbes. 2024. PMID: 38166610 Free PMC article. Review.
-
Transcriptional Profiles of a Foliar Fungal Endophyte (Pestalotiopsis, Ascomycota) and Its Bacterial Symbiont (Luteibacter, Gammaproteobacteria) Reveal Sulfur Exchange and Growth Regulation during Early Phases of Symbiotic Interaction.mSystems. 2022 Apr 26;7(2):e0009122. doi: 10.1128/msystems.00091-22. Epub 2022 Mar 16. mSystems. 2022. PMID: 35293790 Free PMC article.
-
Current knowledge of the Southern Hemisphere marine microbiome in eukaryotic hosts and the Strait of Magellan surface microbiome project.PeerJ. 2023 Oct 3;11:e15978. doi: 10.7717/peerj.15978. eCollection 2023. PeerJ. 2023. PMID: 37810788 Free PMC article. Review.
Cited by
-
Genomic insights reveal community structure and phylogenetic associations of endohyphal bacteria and viruses in fungal endophytes.Environ Microbiome. 2025 Jul 25;20(1):95. doi: 10.1186/s40793-025-00757-8. Environ Microbiome. 2025. PMID: 40713930 Free PMC article.
-
Genetic Characterisation of the Bacterial Microbiota Associating With a Strain of Epichloë Fungal Endophyte of Perennial Ryegrass and the Interaction With Its Paenibacillus Members.Environ Microbiol Rep. 2025 Jun;17(3):e70113. doi: 10.1111/1758-2229.70113. Environ Microbiol Rep. 2025. PMID: 40485104 Free PMC article.
-
Hidden Allies: Decoding the Core Endohyphal Bacteriome of Aspergillus fumigatus.Environ Microbiol Rep. 2025 Aug;17(4):e70153. doi: 10.1111/1758-2229.70153. Environ Microbiol Rep. 2025. PMID: 40831189 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

