Anti-Tumor Effect of Turandot Proteins Induced via the JAK/STAT Pathway in the mxc Hematopoietic Tumor Mutant in Drosophila
- PMID: 37626857
- PMCID: PMC10453024
- DOI: 10.3390/cells12162047
Anti-Tumor Effect of Turandot Proteins Induced via the JAK/STAT Pathway in the mxc Hematopoietic Tumor Mutant in Drosophila
Abstract
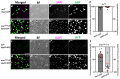
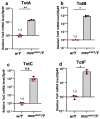
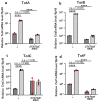
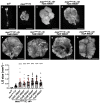
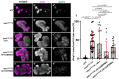
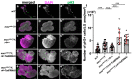
Several antimicrobial peptides suppress the growth of lymph gland (LG) tumors in Drosophila multi sex comb (mxc) mutant larvae. The activity of another family of polypeptides, called Turandots, is also induced via the JAK/STAT pathway after bacterial infection; however, their influence on Drosophila tumors remains unclear. The JAK/STAT pathway was activated in LG tumors, fat body, and circulating hemocytes of mutant larvae. The mRNA levels of Turandot (Tot) genes increased markedly in the mutant fat body and declined upon silencing Stat92E in the fat body, indicating the involvement of the JAK/STAT pathway. Furthermore, significantly enhanced tumor growth upon a fat-body-specific silencing of the mRNAs demonstrated the antitumor effects of these proteins. The proteins were found to be incorporated into small vesicles in mutant circulating hemocytes (as previously reported for several antimicrobial peptides) but not normal cells. In addition, more hemocytes containing these proteins were found to be associated with tumors. The mutant LGs contained activated effector caspases, and a fat-body-specific silencing of Tots inhibited apoptosis and increased the number of mitotic cells in the LG, thereby suggesting that the proteins inhibited tumor cell proliferation. Thus, Tot proteins possibly exhibit antitumor effects via the induction of apoptosis and inhibition of cell proliferation.
Keywords: JAK/STAT; Turandots; antitumor; apoptosis; drosophila; mxc.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Anti-Tumor Effects of Cecropin A and Drosocin Incorporated into Macrophage-like Cells Against Hematopoietic Tumors in Drosophila mxc Mutants.Cells. 2025 Mar 7;14(6):389. doi: 10.3390/cells14060389. Cells. 2025. PMID: 40136638 Free PMC article.
-
Macrophage-like Blood Cells Are Involved in Inter-Tissue Communication to Activate JAK/STAT Signaling, Inducing Antitumor Turandot Proteins in Drosophila Fat Body via the TNF-JNK Pathway.Int J Mol Sci. 2024 Dec 6;25(23):13110. doi: 10.3390/ijms252313110. Int J Mol Sci. 2024. PMID: 39684820 Free PMC article.
-
Loss of Histone Locus Bodies in the Mature Hemocytes of Larval Lymph Gland Result in Hyperplasia of the Tissue in mxc Mutants of Drosophila.Int J Mol Sci. 2020 Feb 26;21(5):1586. doi: 10.3390/ijms21051586. Int J Mol Sci. 2020. PMID: 32111032 Free PMC article.
-
The roles of JAK/STAT signaling in Drosophila immune responses.Immunol Rev. 2004 Apr;198:72-82. doi: 10.1111/j.0105-2896.2004.0133.x. Immunol Rev. 2004. PMID: 15199955 Review.
-
Organogenesis and tumorigenesis: insight from the JAK/STAT pathway in the Drosophila eye.Dev Dyn. 2010 Oct;239(10):2522-33. doi: 10.1002/dvdy.22394. Dev Dyn. 2010. PMID: 20737505 Free PMC article. Review.
Cited by
-
Dual effects of Camellia sinensis and Andrographis paniculata on hyperglycemia and infection in Drosophila.Narra J. 2025 Apr;5(1):e1972. doi: 10.52225/narra.v5i1.1972. Epub 2024 Feb 12. Narra J. 2025. PMID: 40352246 Free PMC article.
-
Anti-Tumor Effects of Cecropin A and Drosocin Incorporated into Macrophage-like Cells Against Hematopoietic Tumors in Drosophila mxc Mutants.Cells. 2025 Mar 7;14(6):389. doi: 10.3390/cells14060389. Cells. 2025. PMID: 40136638 Free PMC article.
-
Activation of immune defences against parasitoid wasps does not underlie the cost of infection.Front Immunol. 2023 Dec 7;14:1275923. doi: 10.3389/fimmu.2023.1275923. eCollection 2023. Front Immunol. 2023. PMID: 38130722 Free PMC article.
-
The potential link between the development of Alzheimer's disease and osteoporosis.Biogerontology. 2025 Jan 20;26(1):43. doi: 10.1007/s10522-024-10181-z. Biogerontology. 2025. PMID: 39832071 Free PMC article. Review.
-
Integrative in silico and in vivo Drosophila model studies reveal the anti-inflammatory, antioxidant, and anticancer properties of red radish microgreen extract.Sci Rep. 2025 May 27;15(1):18533. doi: 10.1038/s41598-025-02999-5. Sci Rep. 2025. PMID: 40425671 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

