The Long Non-Coding RNA ANRIL in Cancers
- PMID: 37627188
- PMCID: PMC10453084
- DOI: 10.3390/cancers15164160
The Long Non-Coding RNA ANRIL in Cancers
Abstract
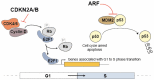
ANRIL (Antisense Noncoding RNA in the INK4 Locus), a long non-coding RNA encoded in the human chromosome 9p21 region, is a critical factor for regulating gene expression by interacting with multiple proteins and miRNAs. It has been found to play important roles in various cellular processes, including cell cycle control and proliferation. Dysregulation of ANRIL has been associated with several diseases like cancers and cardiovascular diseases, for instance. Understanding the oncogenic role of ANRIL and its potential as a diagnostic and prognostic biomarker in cancer is crucial. This review provides insights into the regulatory mechanisms and oncogenic significance of the 9p21 locus and ANRIL in cancer.
Keywords: 9p21 locus; ANRIL; LncRNA; cancer; competitive endogenous RNA; epigenetics; gene regulation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Long Noncoding RNA ANRIL: Lnc-ing Genetic Variation at the Chromosome 9p21 Locus to Molecular Mechanisms of Atherosclerosis.Front Cardiovasc Med. 2018 Nov 6;5:145. doi: 10.3389/fcvm.2018.00145. eCollection 2018. Front Cardiovasc Med. 2018. PMID: 30460243 Free PMC article. Review.
-
Down-regulation of long non-coding RNA antisense non-coding RNA in the INK4 locus suppresses OVCAR-3 cells proliferation and induction of apoptosis by Wnt/β-catenin.J Pharm Pharmacol. 2021 Aug 12;73(9):1212-1217. doi: 10.1093/jpp/rgab042. J Pharm Pharmacol. 2021. PMID: 33772549
-
Potential Links between ANRIL and MiRNAs in Various Cancers.Comb Chem High Throughput Screen. 2024 Jun 7. doi: 10.2174/0113862073294838240523035706. Online ahead of print. Comb Chem High Throughput Screen. 2024. PMID: 38859774
-
Alu elements in ANRIL non-coding RNA at chromosome 9p21 modulate atherogenic cell functions through trans-regulation of gene networks.PLoS Genet. 2013;9(7):e1003588. doi: 10.1371/journal.pgen.1003588. Epub 2013 Jul 4. PLoS Genet. 2013. PMID: 23861667 Free PMC article.
-
Long non-coding RNA ANRIL in gene regulation and its duality in atherosclerosis.J Huazhong Univ Sci Technolog Med Sci. 2017 Dec;37(6):816-822. doi: 10.1007/s11596-017-1812-y. Epub 2017 Dec 21. J Huazhong Univ Sci Technolog Med Sci. 2017. PMID: 29270737 Review.
Cited by
-
lncRNA Biomarkers of Glioblastoma Multiforme.Biomedicines. 2024 Apr 23;12(5):932. doi: 10.3390/biomedicines12050932. Biomedicines. 2024. PMID: 38790894 Free PMC article. Review.
-
LncRNAs as potential prognosis/diagnosis markers and factors driving drug resistance of osteosarcoma, a review.Front Endocrinol (Lausanne). 2024 Jul 2;15:1415722. doi: 10.3389/fendo.2024.1415722. eCollection 2024. Front Endocrinol (Lausanne). 2024. PMID: 39015175 Free PMC article. Review.
-
Disease-Associated Risk Variants and Expression Levels of the lncRNA, CDKN2B-AS1, Are Associated With the Progression of HCC.J Cell Mol Med. 2025 Mar;29(6):e70496. doi: 10.1111/jcmm.70496. J Cell Mol Med. 2025. PMID: 40105653 Free PMC article.
-
Interactions between long non-coding RNAs and m6 A modification in cancer.Discov Oncol. 2025 Apr 20;16(1):579. doi: 10.1007/s12672-025-02387-5. Discov Oncol. 2025. PMID: 40253659 Free PMC article. Review.
-
The role of notch signaling pathway and non-coding RNAs in cancer and inflammation: progress, therapeutic insights, and future directions.Front Immunol. 2025 Jun 20;16:1567040. doi: 10.3389/fimmu.2025.1567040. eCollection 2025. Front Immunol. 2025. PMID: 40621472 Free PMC article. Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

