Rapid Classification of Sarcomas Using Methylation Fingerprint: A Pilot Study
- PMID: 37627196
- PMCID: PMC10453223
- DOI: 10.3390/cancers15164168
Rapid Classification of Sarcomas Using Methylation Fingerprint: A Pilot Study
Abstract
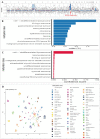
Sarcoma classification is challenging and can lead to treatment delays. Previous studies used DNA aberrations and machine-learning classifiers based on methylation profiles for diagnosis. We aimed to classify sarcomas by analyzing methylation signatures obtained from low-coverage whole-genome sequencing, which also identifies copy-number alterations. DNA was extracted from 23 suspected sarcoma samples and sequenced on an Oxford Nanopore sequencer. The methylation-based classifier, applied in the nanoDx pipeline, was customized using a reference set based on processed Illumina-based methylation data. Classification analysis utilized the Random Forest algorithm and t-distributed stochastic neighbor embedding, while copy-number alterations were detected using a designated R package. Out of the 23 samples encompassing a restricted range of sarcoma types, 20 were successfully sequenced, but two did not contain tumor tissue, according to the pathologist. Among the 18 tumor samples, 14 were classified as reported in the pathology results. Four classifications were discordant with the pathological report, with one compatible and three showing discrepancies. Improving tissue handling, DNA extraction methods, and detecting point mutations and translocations could enhance accuracy. We envision that rapid, accurate, point-of-care sarcoma classification using nanopore sequencing could be achieved through additional validation in a diverse tumor cohort and the integration of methylation-based classification and other DNA aberrations.
Keywords: classification; copy-number; machine learning; methylation; nanopore; sarcoma.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Amin M.B., Greene F.L., Edge S.B., Compton C.C., Gershenwald J.E., Brookland R.K., Meyer L., Gress D.M., Byrd D.R., Winchester D.P. The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J. Clin. 2017;67:93–99. doi: 10.3322/caac.21388. - DOI - PubMed
-
- den Hollander D., Van der Graaf W.T.A., Fiore M., Kasper B., Singer S., Desar I.M.E., Husson O. Unravelling the heterogeneity of soft tissue and bone sarcoma patients’ health-related quality of life: A systematic literature review with focus on tumour location. ESMO Open. 2020;5:e000914. doi: 10.1136/esmoopen-2020-000914. - DOI - PMC - PubMed
-
- Martinez-Trufero J., Cruz Jurado J., Gomez-Mateo M.C., Bernabeu D., Floria L.J., Lavernia J., Sebio A., Garcia Del Muro X., Alvarez R., Correa R., et al. Uncommon and peculiar soft tissue sarcomas: Multidisciplinary review and practical recommendations for diagnosis and treatment. Spanish group for Sarcoma research (GEIS—GROUP). Part I. Cancer Treat. Rev. 2021;99:102259. doi: 10.1016/j.ctrv.2021.102259. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

