The Role of Poly(ADP-ribose) Polymerase 1 in Nuclear and Mitochondrial Base Excision Repair
- PMID: 37627260
- PMCID: PMC10452840
- DOI: 10.3390/biom13081195
The Role of Poly(ADP-ribose) Polymerase 1 in Nuclear and Mitochondrial Base Excision Repair
Abstract
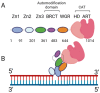
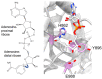
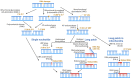
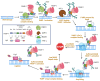
Poly(ADP-ribose) (PAR) Polymerase 1 (PARP-1), also known as ADP-ribosyl transferase with diphtheria toxin homology 1 (ARTD-1), is a critical player in DNA damage repair, during which it catalyzes the ADP ribosylation of self and target enzymes. While the nuclear localization of PARP-1 has been well established, recent studies also suggest its mitochondrial localization. In this review, we summarize the differences between mitochondrial and nuclear Base Excision Repair (BER) pathways, the involvement of PARP-1 in mitochondrial and nuclear BER, and its functional interplay with other BER enzymes.
Keywords: DNA repair; PARP-1; nuclear and mitochondrial localization.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
p21CDKN1A Regulates the Binding of Poly(ADP-Ribose) Polymerase-1 to DNA Repair Intermediates.PLoS One. 2016 Jan 5;11(1):e0146031. doi: 10.1371/journal.pone.0146031. eCollection 2016. PLoS One. 2016. PMID: 26730949 Free PMC article.
-
Quantitative Analysis of Nuclear Poly(ADP-Ribose) Dynamics in Response to Laser-Induced DNA Damage.Methods Mol Biol. 2023;2609:43-59. doi: 10.1007/978-1-0716-2891-1_3. Methods Mol Biol. 2023. PMID: 36515828 Free PMC article.
-
Poly(ADP-ribose) contributes to an association between poly(ADP-ribose) polymerase-1 and xeroderma pigmentosum complementation group A in nucleotide excision repair.J Biol Chem. 2012 Nov 16;287(47):39824-33. doi: 10.1074/jbc.M112.393504. Epub 2012 Oct 4. J Biol Chem. 2012. PMID: 23038248 Free PMC article.
-
Poly(ADP-ribose) Polymerase (PARP) and PARP Inhibitors: Mechanisms of Action and Role in Cardiovascular Disorders.Cardiovasc Toxicol. 2018 Dec;18(6):493-506. doi: 10.1007/s12012-018-9462-2. Cardiovasc Toxicol. 2018. PMID: 29968072 Review.
-
Poly-ADP ribosylation in DNA damage response and cancer therapy.Mutat Res Rev Mutat Res. 2019 Apr-Jun;780:82-91. doi: 10.1016/j.mrrev.2017.09.004. Epub 2017 Sep 20. Mutat Res Rev Mutat Res. 2019. PMID: 31395352 Free PMC article. Review.
Cited by
-
Anoikis-related biomarkers PARP1 and SDCBP as diagnostic and therapeutic targets for asthma.Sci Rep. 2025 Jul 9;15(1):24779. doi: 10.1038/s41598-025-09979-9. Sci Rep. 2025. PMID: 40634444 Free PMC article.
-
Ivosidenib Confers BRCAness Phenotype and Synthetic Lethality to Poly (ADP-Ribose) Polymerase Inhibition in BRCA1/2-Proficient Cancer Cells.Biomedicines. 2025 Apr 14;13(4):958. doi: 10.3390/biomedicines13040958. Biomedicines. 2025. PMID: 40299557 Free PMC article.
-
Protein-Protein Interactions in Base Excision Repair.Biomolecules. 2025 Jun 18;15(6):890. doi: 10.3390/biom15060890. Biomolecules. 2025. PMID: 40563529 Free PMC article. Review.
-
Emerging treatment approaches for triple-negative breast cancer.Med Oncol. 2023 Dec 1;41(1):5. doi: 10.1007/s12032-023-02257-6. Med Oncol. 2023. PMID: 38038783 Review.
-
Non-Neovascular Age-Related Macular Degeneration Assessment: Focus on Optical Coherence Tomography Biomarkers.Diagnostics (Basel). 2024 Apr 3;14(7):764. doi: 10.3390/diagnostics14070764. Diagnostics (Basel). 2024. PMID: 38611677 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

