Divergent Pharmacology and Biased Signaling of the Four Melanocortin-4 Receptor Isoforms in Rainbow Trout (Oncorhynchus mykiss)
- PMID: 37627313
- PMCID: PMC10452266
- DOI: 10.3390/biom13081248
Divergent Pharmacology and Biased Signaling of the Four Melanocortin-4 Receptor Isoforms in Rainbow Trout (Oncorhynchus mykiss)
Abstract
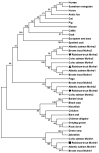
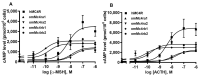
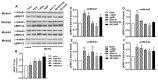
The melanocortin-4 receptor (MC4R) is essential for the modulation of energy balance and reproduction in both fish and mammals. Rainbow trout (Oncorhynchus mykiss) has been extensively studied in various fields and provides a unique opportunity to investigate divergent physiological roles of paralogues. Herein we identified four trout mc4r (mc4ra1, mc4ra2, mc4rb1, and mc4rb2) genes. Four trout Mc4rs (omMc4rs) were homologous to those of teleost and mammalian MC4Rs. Multiple sequence alignments, a phylogenetic tree, chromosomal synteny analyses, and pharmacological studies showed that trout mc4r genes may have undergone different evolutionary processes. All four trout Mc4rs bound to two peptide agonists and elevated intracellular cAMP levels dose-dependently. High basal cAMP levels were observed at two omMc4rs, which were decreased by Agouti-related peptide. Only omMc4rb2 was constitutively active in the ERK1/2 signaling pathway. Ipsen 5i, ML00253764, and MCL0020 were biased allosteric modulators of omMc4rb1 with selective activation upon ERK1/2 signaling. ML00253764 behaved as an allosteric agonist in Gs-cAMP signaling of omMc4rb2. This study will lay the foundation for future physiological studies of various mc4r paralogs and reveal the evolution of MC4R in vertebrates.
Keywords: MC4R; allosteric ligand; biased signaling; constitutive activity; paralogs; rainbow trout.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

