Integrated Multi-Omics Analysis for Inferring Molecular Players in Inclusion Body Myositis
- PMID: 37627634
- PMCID: PMC10452026
- DOI: 10.3390/antiox12081639
Integrated Multi-Omics Analysis for Inferring Molecular Players in Inclusion Body Myositis
Abstract
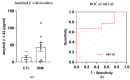
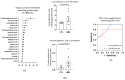
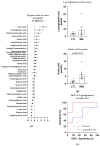
Inclusion body myositis (IBM) is an acquired inflammatory myopathy affecting proximal and distal muscles that leads to weakness in patients over 50. It is diagnosed based on clinical and histological findings in muscle related to inflammation, degeneration, and mitochondria. In relation to IBM, a shortage of validated disease models and a lack of biomarkers and effective treatments constitute an unmet medical need. To overcome these hurdles, we performed an omics analysis of multiple samples from IBM patients (saliva, fibroblasts, urine, plasma, and muscle) to gain insight into the pathophysiology of IBM. Degeneration was evident due to the presence of amyloid β peptide 1-42 (Aβ1-42) in the saliva of the analyzed IBM patients. The presence of metabolic disarrangements in IBM was indicated by an imbalanced organic acid profile in fibroblasts and urine. Specifically, abnormal levels of L-pyroglutamic and orotic acid were supported by the abnormal expression of related metabolites in plasma and urine (glutathione and pyrimidines) and the aberrant expression of upstream gene regulators (L2HGDH, IDH2, OPLAH, and ASL) in muscle. Combined levels of L-pyroglutamic and orotic acid displayed an outstanding biomarker signature in urine with 100% sensitivity and specificity. The confirmation of systemic metabolic disarrangements in IBM and the identification of novel biomarkers reported herein unveil novel insights that require validation in larger cohorts.
Keywords: biomarker; inclusion body myositis (IBM); metabolism; nucleotides; organic acids.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures




References
-
- Buzkova J., Nikkanen J., Ahola S., Hakonen A.H., Sevastianova K., Hovinen T., Yki-Järvinen H., Pietiläinen K.H., Lönnqvist T., Velagapudi V., et al. Metabolomes of Mitochondrial Diseases and Inclusion Body Myositis Patients: Treatment Targets and Biomarkers. EMBO Mol. Med. 2018;10:9091. doi: 10.15252/emmm.201809091. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

